1. Introduction
The field of metabolic engineering has witnessed rapid advancements, further consolidating it as an enabling technology for engineering biological cell factories for producing value-added chemicals and bio-products. The three pillars of metabolic pathway engineering are to achieve high titers, yield and productivity of desired product without detrimental effects on cell growth. Hence, it is critical to develop metabolite-based biosensors to execute feedback control and decouple metabolite production from cell growth. A recent crucial contribution by synthetic biology to engineer more efficient microbial cell factory is the development of transcriptional factor (TF)-based biosensors. Whether coupled with a readable output to enable high-throughput screening or integrated into a genetic circuit to dynamically regulate key metabolic pathways, TF-based biosensors are an indispensable tool for redistributing carbon flux and adapt cell metabolism to the changing environment (Liao and Oh, 1999, Mainguet and Liao, 2010, van der Meer and Belkin, 2010). Such synthetic biosensors are derived from naturally evolved transcriptional factors that propagate changing environmental signals or cellular status into a transcriptional output or cellular phenotype that promotes either cell viability, survival or metabolic economics (Harrison and Dunlop, 2012, Liu et al., 2015). Structural feature of this biosensor is generally divided into two parts: a metabolite-responsive transcriptional regulator and a fluorescence-coupled or fitness-related read-out module (Harrison and Dunlop, 2012, Rogers et al., 2015). This enables metabolic engineers to efficiently quantify varying concentration of cellular metabolites in contrast to the laborious, time-consuming and low throughput analytical methods such as HPLC and LC-MS (Dietrich et al., 2013, Liu et al., 2015). Metabolite-sensing genetic circuits have been reported for sensing various metabolites including macrolides (Mohrle et al., 2007), acetyl phosphate (Farmer and Liao, 2000), farnesyl pyrophosphate(Dahl et al., 2013), 3-hydroxypropanoic acid (Rogers et al., 2015, Rogers and Church, 2016), 1-butanol (Dietrich et al., 2013), and more recently, malonyl-CoA (Ellis and Wolfgang, 2012).
The use of biosensors for in vivo detection and/or quantification of metabolites essentially creates an input-output communication platform within biological cells. This platform has predominantly been exploited to monitor metabolite levels in real-time, especially for the detection of accumulated intermediates or metabolites present in relatively low abundance. Access to such crucial information allows metabolic engineers to gain a deep understanding of kinetics and regulatory mechanisms underlying the engineered metabolic network. In turn, this enables the design of more robust and effective metabolic intervention strategies to maximize production titers of metabolites of interest. That said, biosensors have also found front-end applications as dynamic metabolic pathway regulators and back-end applications as screening devices. When applied to dynamic metabolic pathway regulation, biosensors allow the cell to probe the exact metabolic state and actuate pathway expression that compensates for the metabolic activity of the engineered pathway, and improves the overall productivity and fitness of the engineered cell factory (Xu et al., 2014b). Biosensors can also be developed into a high-throughput screening platform by coupling with a readable output such as fluorescence. This approach is often used to select for high-producing genetic variants or to identify process conditions leading to high product titers (Dietrich et al., 2010, Dietrich et al., 2013, Williams et al., 2016). These applications potentially help to accelerate design-build-test cycles in the engineering of metabolic pathways by facilitating genotype manipulation-phenotype evaluation processes (Rogers et al., 2015, Rogers and Church, 2016).
Over all, many of these applications require that such sensors are orthogonal (without sensor cross-talk) and tunable, allowing for customized biosensor output relative to expected concentrations of metabolites for a range of physiological conditions. This constraint could be decomposed into three pillars: specificity, sensitivity and dynamic range, which are the primary design considerations for their proper function inside the cell. In this review, we explore the fundamental design principles and strategies in modifying and tuning the metabolite-responsive transcription factor (MRTF), using malonyl-CoA biosensor as a prototype. We also explore how these genetically encoded circuits have been successfully applied as a tool in metabolic engineering to dynamically regulate intracellular malonyl-CoA metabolite pools and improve production of malonyl-CoA-derived chemicals in notable host microorganisms.
2. Malonyl-CoA – a vital metabolite
In practically every living system, a portion of the acetyl-CoA flux from the central metabolic pathway is diverted to malonyl-CoA for fatty acid and lipid membrane synthesis, with the aid of acetyl-CoA carboxylase (ACC). This suggests the vital roles that malonyl-CoA plays in cell metabolism and structure. Specifically, malonyl-CoA is a rate limiting substrate for fatty acid synthesis, which in turn, is pivotal for maintaining cell membrane integrity and energy conservation (Schujman et al., 2003, Schujman et al., 2006, Schujman et al., 2008). In mammals, malonyl-CoA has been identified as a crucial fatty acid oxidation regulator which inhibits the mitochondrial carnitine palmitoyltransferase (CPT) − an enzyme involved in fatty acid uptake in the heart and skeletal muscle (Folmes and Lopaschuk, 2007, Foster, 2012). This makes it an effective therapeutic target molecule for treating diseases caused by poor or excessive fatty acid uptake. This has also attracted medical interest in drug development to control malonyl-CoA metabolism at the enzymatic level (Folmes and Lopaschuk, 2007). More relevant to this review, malonyl-CoA is the signaling molecule for regulating lipid metabolism in many Gram-positive bacteria, making it a desirable therapeutic target (Nunn et al., 1977, Dirusso et al., 1993, Magnuson et al., 1993, Marrakchi et al., 2001, James and Cronan, 2003, Schujman et al., 2003, Schujman et al., 2006, Schujman et al., 2008, Yao et al., 2012).
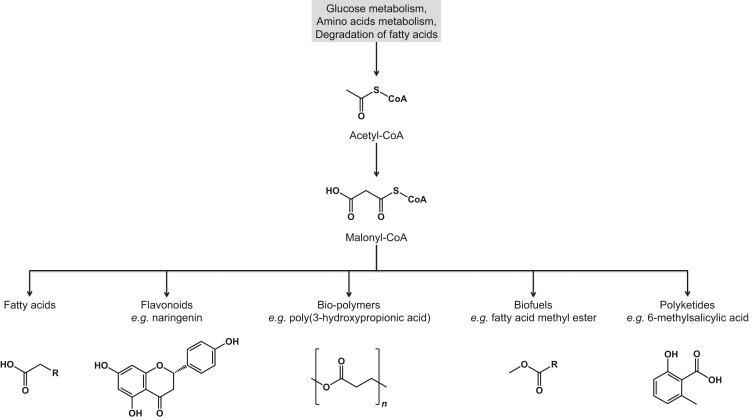
In addition, malonyl-CoA is the building block for many commercially viable bio-products, such as biofuels, biopolymers, and plant natural products. Hence, it has garnered significant interest from the industrial biotechnology sector as a key metabolite target in their engineering of microbial cell factories, (James and Cronan, 2003, Schujman et al., 2003, Schujman et al., 2006) (Fig. 1). These end chemicals are potentially useful as pharmaceutical intermediates, biofuels or other potentially useful chemical products vital to a sustainable bio-economy (Xu et al., 2014a, Xu et al., 2014b, Liu et al., 2015).
 Fig. 1. Compounds derived from malonyl-CoA. Malonyl-CoA, a direct product of acetyl-CoA, can be used as a precursor for the synthesis of fatty acids, flavonoids, bio-polymers, biofuels, and polyketides.
Fig. 1. Compounds derived from malonyl-CoA. Malonyl-CoA, a direct product of acetyl-CoA, can be used as a precursor for the synthesis of fatty acids, flavonoids, bio-polymers, biofuels, and polyketides.However, a major challenge in malonyl-CoA pathway engineering is the perpetually low intracellular concentrations of malonyl-CoA in microbial hosts (4–90 μM or 0.01–0.23 nmol/mg dry weight in E. coli), thus necessitating the use of various metabolic engineering approaches to realize its commercial-scale applications (Takamura et al., 1985, Takamura and Nomura, 1988, Miyahisa et al., 2005). Early metabolic engineering strategies employed static manipulation of relevant pathway enzymes that are directly or indirectly involved in malonyl-CoA metabolism in conjunction with metabolic pathways to channel malonyl-CoA flux to the synthesis of desired products. Some of these approaches include overexpression of acetyl-CoA carboxylase (ACC) – the enzyme that converts acetyl-CoA to malonyl-CoA, increasing intracellular availability of acetyl-CoA – a malonyl-CoA precursor, and down-regulating malonyl-CoA sink pathways (Fig. 2 and Table 1). As such, early attempts at malonyl-CoA engineering relied on conventional analytical techniques such as HPLC and LC to quantify increased concentrations of malonyl-CoA and the desired end product in order to validate the effectiveness of various engineering strategies.
 Fig. 2. A summary of metabolic engineering strategies to increase intracellular malonyl-CoA concentration through rational design. Increasing the pool of acetyl-CoA, decreasing the flux of acetyl-CoA towards non-malonyl-CoA producing acetyl-CoA sink pathways, increasing the expression and/or activity of acetyl-CoA carboxylase and decreasing the flux of malonyl-CoA towards fatty acid synthesis, are direct ways of improving intracellular malonyl-CoA accumulation.
Fig. 2. A summary of metabolic engineering strategies to increase intracellular malonyl-CoA concentration through rational design. Increasing the pool of acetyl-CoA, decreasing the flux of acetyl-CoA towards non-malonyl-CoA producing acetyl-CoA sink pathways, increasing the expression and/or activity of acetyl-CoA carboxylase and decreasing the flux of malonyl-CoA towards fatty acid synthesis, are direct ways of improving intracellular malonyl-CoA accumulation.Table 1. Notable examples of conventional metabolic engineering strategies to increase intracellular malonyl-CoA amount and end chemicals.
| Strategy | Mechanism | Microbial host | Malonyl-CoA | End chemical | Ref. |
|---|---|---|---|---|---|
| Increased expression of acetyl-CoA carboxylase (ACC) | Overexpressing the four subunits of E. coli acetyl-CoA carboxylase (ACC) in a low copy number plasmid under the control of T7 promoter. | E. coli | 100-fold increase relative to WT | 6-fold increase in free fatty acid synthesis | Davis et al. (2000) |
| Overexpression of the two subunits of Corynebacterium glutamicum acetyl-CoA carboxylase (ACC) in a high copy number plasmid under the control of T7 promoter. | E. coli | Not quantified | 60 mg/L yield in flavanones | Miyahisa et al. (2005) | |
| Changing the promoter of ACC1 to a strong constitutive promoter TEF1. | S. cerevisiae | Not quantified | 60% increase in 6-methyl acetyl–salicylic acid | Wattanachaisaereekul et al. (2008) | |
| Increased activity and expression of native acetyl-CoA carboxylase (ACC) | Site-directed mutagenesis of S659 and A1157 to reduce SNF1-mediated phosphorylation of Acc1; overexpression of Acc1 WT and mutants. | S. cerevisiae | Not quantified | 65% increase in total fatty acid content, 3-fold increase in fatty acid ethyl esters (FAEE), and 3.5-fold increase in 3-hydroxypropionic acid (3-HP) | Shi et al. (2014) |
| Increased acetyl-CoA flux towards malonyl-CoA accumulation | T7 promoter-controlled overexpression of acetyl-CoA carboxylase (ACC), overexpression of acetyl-CoA synthetase, which recycles acetate to acetyl CoA; double knock-out of genes encoding phosphotransacetylase (Pta) and acetate kinase (AckA), which are genes responsible for acetate biosynthesis from acetyl-CoA; deletion of alcohol/aldehyde dehydrogenase (adhE). | E. coli | 15-fold increase relative to WT | 4-fold increase in phloroglucinol production | Zha et al. (2009) |
| Overexpression of the four subunits of Photorhabdus luminescens acetyl-CoA carboxylase (ACC) in a low copy number plasmid and overexpression of Photorhabdus luminescens biotin ligase (BirA) in a high copy number episomal vector. Biotin ligase catalyzes the biotinylation of the biotin-dependent BCCP subunit of ACC. | E. coli | Not quantified | 1166% increase in flavonone synthesis | Leonard et al. (2007) | |
| Decreased expression of malonyl-CoA-consuming enzymes | Use of anti-sense RNA to silence the expression of malonyl-CoA transacylase (FabD) – a fatty acid synthesis enzyme. | E. coli | 4.5-fold increase relative to WT | 2.53-, 1.70-, 1.53-fold increase in the production of 4-hydroxycoumarin, resveratrol, and naringenin, respectively | Yang et al. (2015) |
| Expression of another malonyl-CoA source pathway | Overexpressing the malonate carrier protein (matC), which transports malonate into the cell and malonyl-CoA synthetase (matB), which converts malonate to malonyl-CoA. | E. coli | Not quantified | 100.64 mg/L of (2S)-naringenin | Wu et al. (2014) |
| Use predictive computational models to select a combination of metabolic engineering approaches for strain optimization | Overexpression of acetyl-CoA carboxylase (ACC), phosphoglycerate kinase (PGK) and pyruvate dehydrogenase (PDH), coupled with double knock-out of fumarase (fumC) and succinyl-CoA synthetase (sucC) genes. | E. coli | 3.7-fold increase relative to WT | 474 mg/L naringenin production | Xu et al. (2011) |
| Overexpression of acetyl-CoA carboxylase (ACC), biotin ligase (BirA) and pantothenate kinase in a strain deficient in citric acid cycle genes (sdhCDABand citE), an amino acid transporter gene (brnQ), and an alcohol dehydrogenase gene (adhE) | E. coli | 2.7-fold increase in relative to WT | 660% increase in naringenin synthesis and 420% increase in eriodictyol synthesis | Fowler et al. (2009) |
In addition, an integrated computational and experimental approach has been employed to predict static genetic intervention targets to increase cellular malonyl-CoA concentration (Fowler et al., 2009, Xu et al., 2011). By studying the genome-scale metabolic network of a biological host, such models nominate target genes for knock out and/or over-expression to improve malonyl-CoA amounts. Besides identifying gene targets directly related to malonyl-CoA metabolism, computational tools have been applied in highlighting potential impact of targeted genes on cell growth. Furthermore, they have been used to explore the potential roles of genes that are indirectly related to malonyl-CoA metabolism. The ability to identify distant pathways that are not directly involved in target metabolism demonstrates the effectiveness of genome-scale computational modeling. Overall, computational tools have been successfully applied in finding the most promising combination of approaches to achieve an improved malonyl-CoA yield. As an example, Xu et al. applied a genome-scale metabolic network model to achieve a balanced precursor distribution between biomass production and the synthesis of naringenin, a flavonoid compound, in E. coli (Xu et al., 2011). Guided by OptForce methodology (Ranganathan et al., 2010), the model predicted over-expression of ACC, phosphoglycerate kinase(PGK) and pyruvate dehydrogenase (PDH), in addition to double knockout of genes for the citric acid cycle enzymes: succinyl-CoA synthetase (sucC) and fumarase (fumC). The combination of these metabolic adjustments resulted in a 3.7-fold increase in malonyl-CoA amount and a 5.6-fold increase in naringenin production at 474 mg/L, compared to the wild-type. Similarly, Fowler et al.demonstrated the use of a computational model known as Cipher of Evolutionary Design (CiED) (Fowler et al., 2009). The tool predicted the knockout of several citric acid cycle genes (sdhCDAB and citE), an amino acid transporter (brnQ) and a pyruvate consumer (alcohol dehydrogenase adhE) for engineering an efficient flavonoid producing E. coli strain. This deletion strain was subsequently used to overexpress ACC, acetyl-CoA synthetase, biotin ligaseand pantothenate kinase to achieve 660% and 420% increase in naringenin and eriotictyol production, respectively. These strategies have also led to the development of a high fatty acid-producing strain in later studies (Xu et al., 2013b). Complementing the strategy adopted by Xu et al. (2011) and Fowler et al. (2009) that relies on static control genetic modifications (i.e., overexpression and/or deletion), CRISPR/dCas9-mediated repression of endogenous targets predicted by OptForce was proven a highly promising strategy to increase flavonoid production (Cress et al., 2015).
Besides the implementation of classical metabolic engineering strategies, chemicals that inhibit fatty acid synthesis, such as cerulenin and triclosan, have been shown to effectively boost intracellular malonyl-CoA levels (Davis et al., 2000). Cerulenin up-regulates intracellular malonyl-CoA amounts by inhibiting β-ketoacyl-acyl carrier protein synthase enzymes (FabB and FabF), which condense malonyl-ACP with acyl-ACP to extend the fatty acid chain by two carbon atoms (Schujman et al., 2006, Schujman et al., 2008). However, the inhibition in fatty acid synthesis under high cerulenin concentrations negatively impacts cell viability. The high cost of cerulenin prohibits its use for commercial-scale fermentation production (Davis et al., 2000). Despite its limited commercial application, experimentation with cerulenin concentrations to mimic increasing malonyl-CoA level was crucial in establishing malonyl-CoA as the effector molecule for regulating fatty acid synthesis in Gram-positive bacterial species (Schujman et al., 2003, Schujman et al., 2006).
3. Mechanism of malonyl-CoA sensing
Malonyl-CoA biosensors are a synthetic mimicry of the native fatty acid biosynthesis transcriptional regulatory circuits, found naturally in many Gram-positive bacteria such as Bacillus subtilis, Staphylococcus aureus, Bacillus anthracis, Listeria monocytogenes and Streptococcus pneumonia (James and Cronan, 2003, Fujita et al., 2007, Xu et al., 2014a, Li et al., 2015, Liu et al., 2015). This naturally occurring regulon consists of an autogenously regulated FapR transcriptional repressor module that is typically located adjacent to a fatty acid synthesis operon, whose expression is driven by a hybrid promoter possessing a 17-bp cis-regulatory fapO-operator (James and Cronan, 2003). FapR mediates the repression of fatty acid synthesis genes via DNA-protein interaction with the fapO-operator sequence using its N-terminal regulatory domain (KD = 0.12 μM) (Xu et al., 2014a). This interaction cascades into a FapR-fapO complex that sterically hinders RNA polymerase proceeding forward and blocks transcription of the downstream fatty acid synthesis genes. The C-terminal ligand-binding domain of FapR is typically a thioestease-like structure that specifically recognizes malonyl-CoA. Intracellular accumulation of malonyl-CoA gradually relieves this FapR-mediated repression through metabolite-protein interaction of malonyl-CoA with the C-terminal ligand-binding domain of FapR (KD = 2.4 μM) (Schujman et al., 2003, Schujman et al., 2006, Ellis and Wolfgang, 2012, Xu et al., 2014a). Binding of malonyl-CoA with the C-terminus of FapR triggers a conformational change at the N-terminus of FapR, which destabilizes the FapR-fapO complex and relieves FapR from interacting with the fapO-operator, thereby permitting interaction of RNA polymerase with the promoter, and thus initiating transcription (Fig. 3).
 Fig. 3. Mechanism of malonyl-CoA sensing. Binding of malonyl-CoA to the C-terminus of FapR cascades into a conformation change at its N-terminus, which destabilizes FapR-fapO interaction. This enables the formation of an RNA polymerase-promoter complex, thus inducing transcription of the downstream fatty acid synthesis gene (FASII).
Fig. 3. Mechanism of malonyl-CoA sensing. Binding of malonyl-CoA to the C-terminus of FapR cascades into a conformation change at its N-terminus, which destabilizes FapR-fapO interaction. This enables the formation of an RNA polymerase-promoter complex, thus inducing transcription of the downstream fatty acid synthesis gene (FASII).4. The architecture of a malonyl-CoA biosensor
The design of a typical malonyl-CoA-sensor (Fig. 4) requires the integration of the malonyl-CoA-responsive transcriptional factor FapR with a hybrid promoter containing the DNA binding site for FapR (Fig. 3). A repressor module is located on one end of the circuitry, comprising a fapR gene and a suitable promoter driving FapR expression. In the reporter module on the other end, there is a reporter gene typically encoding a fluorescence protein (e.g., egfp, rfp, mCherryand tdTomato); the expression of which is driven by a fapO-hybrid promoter with the cis-regulatory fapO-operator sequence located within or adjacent to the reporter promoter. Identical to the naturally occurring regulons, the reporter module translates the increase in intracellular malonyl-CoA concentration (i.e., input) into the expression level of the reporter protein (i.e., output), at a rate that is commensurate with the degree of de-repression of the reporter promoter. Hence, by perturbating the intracellular concentration of malonyl-CoA (e.g., through the use of cerulenin), the resulting increase in fluorescence signal generates a malonyl-CoA concentration-dependent calibration curve. This, in turn, serves as an input-output model for using such malonyl-CoA sensors to quantify intracellular malonyl-CoA concentrations derived from malonyl-CoA source pathways.
 Fig. 4. Schematic diagram of the architecture of a malonyl-CoA sensor showing its modularity with a repressor module (P1 and FapR gene) and a reporter module (P2, fapO and RFP gene). P1 – repressor promoter, P2 – reporter promoter, fapO – fapO operator, T1, T2 – transcriptional terminators.
Fig. 4. Schematic diagram of the architecture of a malonyl-CoA sensor showing its modularity with a repressor module (P1 and FapR gene) and a reporter module (P2, fapO and RFP gene). P1 – repressor promoter, P2 – reporter promoter, fapO – fapO operator, T1, T2 – transcriptional terminators.5. The design criteria of an effective malonyl-CoA biosensor
Engineering an effective malonyl-CoA sensing genetic circuit for a biological host requires a number of design considerations. Firstly, the biological chassis tested (i.e., the host itself) potentially determines the ease of transformation/transfection of the genetic circuit as well as its replication and maintenance within the chassis. This factor must be built into the structural design of the circuit. In other words, the sensor must have a plasmid backbone with an origin of replication that is compatible with the replication machinery of the host (Li et al., 2015). Moreover, a dual plasmid circuit system with separate vectors for the repressor and reporter modules may be required for some organisms, where a large plasmid may prove difficult to be transformed or transfected. This latter case requires additional consideration of plasmid incompatibility and the choice of selection marker. A good example of how these considerations are crucial is demonstrated in the malonyl-CoA biosensors developed to investigate fatty acid metabolism in mammalian cells, where carefully selected genetic parts facilitated sensor functionality, with improved transfection, replication and expression efficiency, in the mammalian host (Ellis and Wolfgang, 2012).
Secondly, a functionally effective sensor requires the proper balance in expression of both the repressor and reporter modules (Fig. 5). This is a pre-requisite for high detection limit and broad dynamic range of response. The upper detection limit of the sensor is determined by how much malonyl-CoA could saturate the FapR repressor (KD constrant), however, also physically constrained by the amount of FapR present inside the cell and how strong FapR interact with the DNA-binding site (fapO). This fact emphasizes the importance to control the amount of the repressor protein FapR. On the other hand, sensitivity is determined by the cooperativity of the FapR-faoO-malonyl-CoA interaction, that said the allosteric effect constant or the Hill coefficient. Increasing the number of FapR-binding site (fapO) might be useful to tune both the detection limit and sensitivity of the engineered malonyl-CoA sensor. Additionally, fine-tuning is necessary to achieve optimal sensitivity (i.e., responsive to small changes in intracellular malonyl-CoA concentration), responsivity (i.e., yielding a measurable read-out value), and a high signal-to-noise ratio in response to malonyl-CoA. This intricate balance requires intracellular FapR repressor concentration to be optimal: high enough to achieve high detection limit, but not too high to avoid poor sensitivity. Likewise, the expression of the reporter module should be high enough to achieve a detectable read-out response, but not too high to avoid detrimental effects on cell growth (Fig. 5).