. Introduction
In research laboratory settings, metabolic engineering has enabled the production of a vast array of metabolite products including fuels (Jiang et al., 2018, 2017; Yan and Pfleger, 2020), chemicals (Bai et al., 2019; Bowen et al., 2016; Reed and Alper, 2018), medicines (Cao et al., 2020), and polymer precursors (Zheng et al., 2020), using dozens of microbial species from bacteria (Becker et al., 2018; Pontrelli et al., 2018) to eukarya (Abdel-Mawgoud et al., 2018; Lian et al., 2018) and archaea domains (Crosby et al., 2019). However, commercial production of these compounds in industrial scales has been lagging, largely due to the inability of the engineered strains to maintain stable performance at large scales while meeting stringent titer, rate, and yield (TRY) requirements.
Metabolic engineers have faced a myriad of challenges in forcing engineered microbes to over-produce metabolite products. Engineered metabolic pathways utilize shared host machinery, including RNA polymerases, ribosomes, ATP, cofactors, and other native metabolites. Competition on cellular resources is sensitive to fermentation conditions and can cause metabolic burden (Kurland and Dong, 1996), improper cofactor balance (Bentley et al., 2016; Charusanti et al., 2010), or accumulation of metabolites to toxic levels (Kizer et al., 2008), all of which can interfere with the growth and desired metabolic objective of the engineered microbes. Additionally, cells growing in large-scale bioreactors often experience different and changing microenvironments, leading to heterogeneity in their performance (Delvigne et al., 2014; Pigou and Morchain, 2015). These issues constrain metabolite production and give advantage to fast-growing, yet non-productive isogenic cells or mutant strains (Rugbjerg et al., 2018a), which ultimately lowers overall TRY performance (Zhuang et al., 2013). Optimizing strain performance through design-build-test cycles is lengthy and costly. The cost of commercializing a metabolite product was estimated to range from $100 million to $1 billion (Crater and Lievense, 2018; Wehrs et al., 2019).
Dynamic metabolic engineering seeks to address these challenges through the development of genetically encoded control systems which allow microbes to autonomously adjust their metabolic flux in response to the external environment and/or internal metabolic state. The concept is inspired by natural metabolic control systems which microbes use to maintain homeostasis, coordinate metabolic flux (Chubukov et al., 2014; Kochanowski et al., 2017), and adapt metabolism to changing environments (Chin et al., 2008) and stresses (Jozefczuk et al., 2010). These dynamic control systems are in contrast to static control systems traditionally used in metabolic engineering, where metabolic pathways are expressed constitutively and are tuned by the choice of promoters, ribosome binding sites, and gene copy numbers (Holtz and Keasling, 2010). Since the first demonstration of enhanced lycopene production two decades ago (Farmer and Liao, 2000), dynamic metabolic engineering has become a popular strategy. Recent advances in synthetic biology and systems biology have provided the tools for dynamic metabolic engineering while control theory has provided new design principles for expanding beyond natural control systems. There are multiple examples where dynamic metabolic control has provided microbes remarkable robustness in different fermentation conditions and improved TRY performance (Anesiadis et al., 2008; Cress et al., 2015; Liu et al., 2018).
In this work, we seek to provide a comprehensive review of the theory and practice of dynamic metabolic engineering, focusing on its improvement to TRY metrics. In the first section, we will present theoretical works that discuss the benefits of incorporating dynamic metabolic control as well as works that provide guidance on design choices for the construction of control systems. In the second section, we will review the construction and engineering of sensors and control mechanisms, referred to as actuators, the two fundamental components of dynamic metabolic control systems. Finally, we will review metabolic pathways to which dynamic metabolic engineering has been applied with the focus on control topologies.
2. Theoretical insights and design choices for metabolic dynamic control
Several design choices need to be made before implementing a dynamic control system in a metabolic pathway. In this section, we summarize the theoretical concepts and benefits of three popular dynamic control strategies: two-stage metabolic switches, continuous metabolic control, and population behavior control. Additionally, for each section we highlight theoretical works which elucidate the design choices for improving TRY metrics.
2.1. Two-stange metabolic control systems
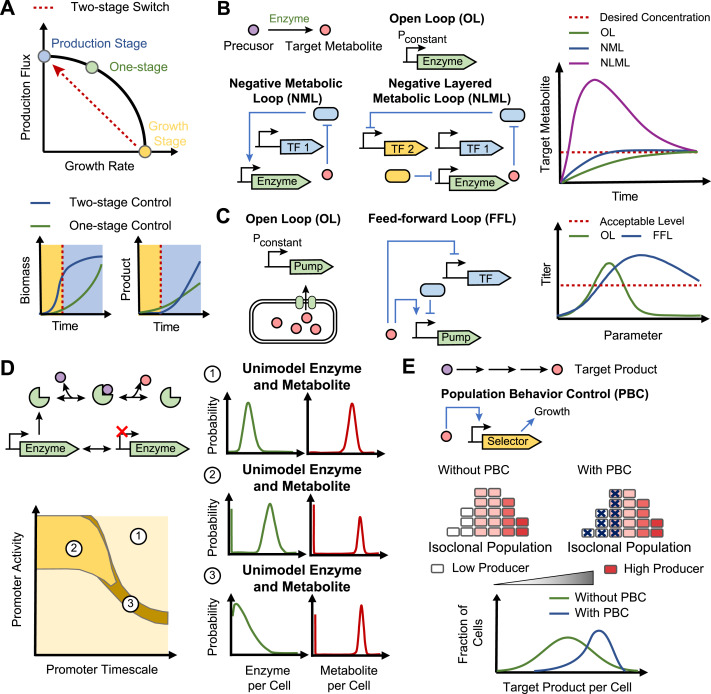
A two-stage metabolic switch is a straightforward yet effective dynamic control strategy for overcoming numerous trade-offs inherent in engineered metabolic bioprocesses (Burg et al., 2016; Venayak et al., 2015). A two-stage metabolic switch decouples the competing tasks of biomass accumulation and metabolite overproduction. In the first stage, cells are engineered to focus on rapid cell growth, usually with minimal production of the product. In the second stage, cell growth is minimized while substrate fluxes are funneled into product formation. This contrasts with a one-stage process where biomass accumulation and product formation occur concurrently. One early example of two-stage process is the batch production of glycerol and ethanol in Escherichia coli (Gadkar et al., 2005). The work found that glycerol concentration could be improved by 30% with a flux switch during production as compared to maintaining constant glycerol flux throughout the batch process (Fig. 1A). In the modeled one-stage production, biomass accumulated more slowly than the optimal two-stage production, leading to slower volumetric productivity and a lower glycerol titer. This demonstrates the value of a two-stage production for optimizing productivity, which can be a critical consideration for commercializing a bioprocess (Zhuang et al., 2013).
 Fig. 1
Fig. 12.1.1. Choosing between a two-stage or one-stage fermentation process
Not all two-stage bioprocesses can outperform one-stage fermentation. Important factors that affect the decision to implement either a one-stage or two-stage bioprocess include strain performance during slow-growing or non-growing conditions, economic constraints (Klamt et al., 2018), as well as metabolic network dynamics (Jabarivelisdeh and Waldherr, 2018). For example, slow-growing and stationary phase cells often have reduced glucose uptake rates which can decrease substrate utilization for product formation (Burg et al., 2016; Harder et al., 2018). This trade-off in substrate utilization and growth was explored by Klamt et al. (2018). The model showed that reducing the glucose uptake rate in the production phase below approximately 4 mmol/gDW/h, which fell in the experimentally observed range of 0.5–4.5 mmol/gDW/h, caused the two-stage process to have lower volumetric productivity than a comparable one-stage process with the same yield and initial conditions.
The mode of bioprocess operation (e.g. fed-batch versus batch) also affects the choice between one-stage and two-stage processes. Yegorov et al. developed a kinetic model to study cellular reactions governing both shared cell resources (e.g. transcription, translation) and bioproduction (e.g. enzymatic reactions in an engineered pathway) (Yegorov et al., 2019). The model was used to find the optimal RNA polymerase expression rate for maximizing either biomass or product formation under constant or limited substrate environments. This work uncovered that in the case of a constant nutritional environment, which can be found in fed-batch and continuous bioprocess, a high RNA polymerase activity to maximize both cell growth and production is preferred, motivating a one-stage fermentation. However, when the nutrient is limited, such as in a batch process, RNA polymerase activity needs to be reduced to shut down cellular replication and focus cellular resources on the expression of product formation enzymes. Thus, batch processes can benefit most from using a two-stage process.
2.1.2. Choosing valves for two-stage switch
To implement a two-stage process, appropriate metabolic reactions must be controlled. Although several well-known strain design algorithms are available for static genetic interventions (Burgard et al., 2003; Ranganathan et al., 2010), these may not be appropriate for two-stage switchable systems. An algorithm for identifying reactions that can be either knocked-out or controlled to achieve near theoretical maximum yield in two metabolic states was developed (Venayak et al., 2018b). This algorithm was applied to identify metabolic valves that can switch from 90% maximum biomass yield to 90% maximum product yield. For 87 organic products that can be derived from E. coli metabolism, 56 of them can be switched using a single switchable valve. A small number of valves in glycolysis, the TCA cycle, and oxidative phosphorylation were found particularly useful for the decoupled production. Thus, this strategy could be highly beneficial for identifying appropriate control valves computationally.
2.1.3. Engineering bistability for two-stage switchable systems
While many biological control systems can be used to switch between two-state, systems which exhibit bistability have additional benefits (Ferrell, 2002; Mitrophanov and Groisman, 2008; Tiwari et al., 2011; Veening et al., 2008). One general property of bistable systems is hysteresis, a memory-like property where the threshold of the signal output response curve is different depending on the recent history of the input signal (Ferrell, 2002). Hysteresis can allow the input signal to be reduced without switching back to the growth state. Additionally, bistability enables a slow response to input signals that are near the switching threshold (Tiwari et al., 2011). This property allows bistable switches to filter out mild, transient changes in the input signal which may be typical of a heterogenous bioreactor. A general design principle for a bistable metabolic system is to have a positive feedback loop along with non-linear kinetics, such as using a cooperative promoter, or ultrasensitive enzyme kinetics (Angeli et al., 2004). One example of bistability in metabolic engineering is the application of a metabolic toggle-switch, which uses mutually repressive transcription factors to lock cells in one of two metabolic states until a metabolic switching signal is applied (Bothfeld et al., 2017; Venayak et al., 2018a). Bistability in metabolic uptake rate could be useful for implementing a two-stage production since the extracellular nutrient concentration could be different in growth and production stage. Several positive feedback architectures for generating bistable metabolite uptake rate were explored (Oyarzún and Chaves, 2015). It was found that using an activator-repressor topology, where the metabolite activates its own uptake and represses its consumption, had the largest parameter space for generating bistability.
2.2. Continuous metabolic control systems
Continuous metabolic control is a dynamic metabolic control strategy where microbes continuously sense and respond to changing signals from its external environment or internal metabolic state. Continuous metabolic control has received increasing attention in recent years (He et al., 2016; Liu et al., 2018) due to its numerous potential benefits to metabolic engineering including automatic balancing of metabolic fluxes (Zhang et al., 2012), alleviating toxicity from intermediate accumulation, accelerating dynamic response (Liu and Zhang, 2018), reducing sensitivity to metabolic perturbation (Oyarzún and Stan, 2013), and making systems robust to parameter uncertainty (Dunlop et al., 2010). An early example of continuous metabolic control is the automatic balancing of free fatty acid (FFA) and ethanol flux in the production of fatty acid ethyl esters (FAEE) in E. coli (Zhang et al., 2012). The FAEE production system had numerous challenges, including ethanol toxicity, competition for metabolite intermediates with native pathways, and futile cycling of FFA activation. The work found that sensing acyl-CoA, a key pathway intermediate, and feedback to the ethanol and FFA producing modules allowed cells to maintain tight control over FAEE production, with a three-fold increase in yield compared to a strain without continuous metabolic control. Additionally, a kinetic model of the FAEE control system revealed that continuous metabolic control of the pathway modules could improve productivity compared to constitutive control over a wide range of parameters. Global sensitivity analysis indicated that the most important parameters are those associated with the accumulation of toxic intermediates and the synthesis of burdensome proteins. Due to the numerous potential benefits, continuous metabolic control could improve the performance of a large range of metabolic pathways.
2.2.1. Choosing valves for continuous dynamic control
Implementing a continuous metabolic control first requires choosing the metabolic reactions to be controlled. Currently, unlike in two-state switching, generally applicable algorithms for identifying the best metabolic reactions for dynamic control have not been developed. The challenge comes from the fact that the problems solved by continuous dynamic control are often pathway-specific, such as to avoid the accumulation of a toxic pathway intermediate or to optimize the expression of a burdensome enzyme. Thus, implementation of continuous dynamic control requires preexisting knowledge on rate-limiting steps, pathway bottlenecks, and intermediate accumulation. Metabolic control analysis (MCA) can potentially be used to identify reactions that have a high degree of control over pathway flux, which would suggest that tight control over these nodes may have the most impact on the pathway (for review of MCA see (He et al., 2016)).
2.2.2. Choosing control topology
Common control topologies in dynamic metabolic engineering are positive feedback, negative feedback, and feedforward. Deciding which topology to use is generally specific to the system and requires a detailed kinetic model to evaluate which topologies will provide the largest gains in TRY metrics. Simulating all possible topologies with many parameters was demonstrated as a method for choosing the best continuous control system (Stevens and Carothers, 2015). In this work, production of p-aminostyrene (p-AS) was simulated for 729 unique possibilities for the control topologies along with 2000 parameter values for each topology. The p-AS yield from each simulation was compared to the median yield from the static control topology to identify topologies which generally improve yields. This work found that only 19% of topologies had higher median yields than static control, however, the top 5% of architecture had median yields of nearly 3 times higher than static control. Although some topologies could generally improve yield, the spread of performance within a topology could be more than 100-fold. In general, there is no topology that will always outperform other topologies in dynamic metabolic control. Thus, choosing the best topology highly depends on the controlled system and would require extensive modeling to uncover the maximum performance.
2.2.3. Accelerating metabolite dynamics through engineered feedback control
In dynamic metabolic systems, rapid response to the switching signal is desired because it allows microbes to reach a productive metabolic state more rapidly, thus potentially improving overall TRY metrics. Incorporating continuous metabolic control into these systems has been recognized as a potential strategy for altering the metabolite dynamics (Schmitz et al., 2017). Several feedback architectures and their parameters have been explored at the theoretical level for accelerating response to metabolic signals.
Metabolic switching using a conventional open loop (OL) pathway topology in response to an input signal, such as induction of enzyme expression, is generally slow. It was found that for an OL topology, it takes 2.48 cell cycles for the metabolite concentration to rise to 50% of its steady-state concentration (defined as the metabolite rise-time) after the step-up input signal (Fig. 1B). Liu et al. built and studied three closed loop feedback systems of different topologies: a negative gene loop (NGL), a negative metabolic loop (NML), and a negative layered metabolic loop (NLML) (Liu and Zhang, 2018) (Fig. 1B). While all three loops had some ability to decrease the rise-time for metabolite concentrations compared to the OL, the NLML, where metabolite concentration is feedback through a genetic inverter then an enzyme controller, was capable of dramatically accelerating rise-times by 11.8-fold (Fig. 1B). However, this rapid increase in rise-time was accompanied by a large metabolite concentration overshoot, where metabolite concentration rises above the steady-state before settling. Through tuning the many parameters, it was found that for NLML, faster rise times were generally correlated with larger overshoots. Using a high maximum promoter strength of the genetic inverter and a low threshold of the enzyme controller were found to be the most efficient for decreasing the rise-time.
In addition to step-up inputs, rapid response to a step-down input signal, or removal of a step-up input signal, could be beneficial in allowing switching systems to recover to their initial states. The recovery dynamics of a metabolite uptake architecture were studied (Hartline et al., 2020). In this architecture, the enzyme for extracellular uptake is controlled by a positive feedback loop where the intracellular metabolite activates the expression of the uptake enzyme by sequestering the metabolite-responsive transcription factor (MRTF) during the step-up response. It was found that once the extracellular metabolite concentration drops, the expression of the uptake enzyme will continue due to the delay in dropping the intracellular metabolite concentration, thus leading to a long time to drop uptake pathway enzyme concentrations to 50% of its initial level, called the recovery time. The recovery dynamics can be accelerated by rapidly releasing the MRTF from sequestration. The rate of release of MRTF depends on the amount of MRTF stored during the high extracellular metabolite state and by the rate of consumption of the intracellular metabolite. MRTF promoter strength was identified as a key parameter for tuning pathway recovery time. Further, incorporating a negative autoregulatory loop could help reduce the overall resource utilization for actuating the recovery dynamics. These modeling results were additionally confirmed through re-engineering and testing of the E. coli fatty acid uptake system.
2.2.4. Improving metabolic robustness through engineered feedback control
In designing a robust metabolically engineered bioprocess, two types of uncertainties should be accounted for: microbial environmental perturbation and parameter uncertainty. Engineering negative feedback in the metabolic control system has been explored as an avenue to make engineered microbes robust to both kinds of uncertainties.
Microbial environmental perturbation arises due to the dynamic and heterogeneous microenvironments that microbes encounter in a bioreactor (Lara et al., 2006). Such perturbations can cause major concentration changes for metabolites involved in an engineered pathway (Kresnowati et al., 2006; Taymaz-Nikerel et al., 2013). Metabolite feedback was shown to help make metabolite levels less sensitive to these metabolic fluctuations (Oyarzún and Stan, 2013, 2012). Using strong, tight promoters allowed the system to further minimize the decrease in product concentration, however, strong promoters could also lead to oscillatory dynamics. Although these theoretical studies are promising, experimental implementation of these control systems have not been performed. Compared to well-controlled lab-scale fermentation, effects from these control systems are probably more dramatic in large-scale fermenters, where microbial environmental uncertainties are more pronounced. Although, metabolic engineers currently focus on lab-scale optimizations, we expect these robust metabolic control systems to demonstrate experimental improvements in the near future.
Parameter uncertainty arises due to the use of biological parts, such as promoters and ribosome binding sites, which have uncertain performance characteristics, and therefore requires extensive fine-tuning to achieve desired system performance. Metabolite feedback has also been explored as a mechanism to make systems less sensitive to uncertainty in system parameters (Dunlop et al., 2010; Harrison and Dunlop, 2012). Toxic biofuel production was modeled as an example system, and the effects of feedback architecture on parameter sensitivity were explored (Dunlop et al., 2010). In this model system, the biofuel product is toxic to cell growth, which hinders further biofuel production. Cellular efflux pumps were modeled to remove biofuel from the cell, reducing the biofuel toxicity. However, over-expression of membrane-bound efflux pumps can be toxic too, thus requiring careful balance of efflux pump expression and biofuel production. For the case of no feedback control in the expression of efflux pumps, the model revealed a narrow range of promoter strengths had a nearly maximal amount of biofuel production (Fig. 1C). Parameter sensitivity analysis of three other controller architectures demonstrated that including feedback in general reduced the sensitivity of biofuel production to system parameters variation. Further, it was found that the feedforward loop topology was robust to parameter uncertainty for all five model parameters (Fig. 1C). Based on this modeling work, an experimental library of feedback systems was constructed and tested for enhanced tolerance to the biofuel pinene (Siu et al., 2018). Several members of this library were shown to have enhanced tolerance to pinene, despite having varying promoter strengths and number of transcription factor binding sites in the promoter. These results highlight how incorporating feedback control into pathway design can make the system more robust to parametric uncertainties.
2.3. Control of metabolite heterogeneity
Metabolite heterogeneity, the cell-to-cell variation in metabolite levels in isogenic populations (Schmitz et al., 2017), has only recently been recognized to strongly affect overall TRY performance (Lv et al., 2019). Microbes have numerous stochastic mechanisms that give rise to cell-to-cell differences in mRNA, proteins, metabolic levels, and flux activities (Ackermann, 2015; Raj and van Oudenaarden, 2008; Takhaveev and Heinemann, 2018). Here, we discuss theoretical insights on the impact of metabolic heterogeneity on metabolic bioprocesses and how dynamic metabolic control can reduce or exploit metabolite heterogeneity for improved bioprocess performance.
2.3.1. Stochasticity in metabolic pathways
Stochasticity in biology can lead to large heterogeneity in metabolic activity. Using experimental measurements of the single-cell proteomic data (Taniguchi et al., 2010), the impact of cell-to-cell variation on protein concentration to E. coli population growth rate has been assessed by a genome-scale FBA model (Labhsetwar et al., 2013). The model produced a population with a broad distribution of growth rates. Cells at the extremes of the growth rate distribution had different metabolic fluxes, where the fast-growing cells tended to prefer using the Entner-Doudoroff pathway, while slow-growing cells utilized the glycolysis pathway more predominantly. These stochastic effects could be important for production, particularly if production is growth-coupled or if the product is derived from a pathway intermediate which varies strongly between cells.
Additionally, large metabolite heterogeneity can arise from enzyme variation even within one metabolic reaction (Tonn et al., 2019). Stochastic analysis of a model metabolic pathway with reversible Michaelis-Menten kinetics revealed several regimes of bimodal metabolite distributions (Fig. 1D). In the first regime, promoters with slow switching but high expression rates generate a bimodal distribution in enzyme levels, leading to bimodality in metabolites. In the second regime, enzymes are unimodally expressed at a low level. Occasionally, enzyme concentration could reach near-zero, which caused the product to become depleted, so a bimodal distribution in metabolite levels was generated (Fig. 1D). In engineered systems, enzymes are typically highly expressed, making it unlikely for the second bimodality regime to occur. However, many products are derived from native metabolites, which could exhibit this mode of bimodality. In these cases, a cell may stochastically have low levels of production pathway substrate, which may impact bioprocess productivity. These theoretical studies have raised the concern that metabolite heterogeneity impacts bioprocess robustness (Delvigne and Goffin, 2014) and thus warrant strategies to cope with heterogeneity (Binder et al., 2017).
2.3.2. Controlling metabolite heterogeneity using feedback loops
Several papers have explored the impact of feedback on the amount of metabolite heterogeneity in the system (Borri et al., 2016, 2015; Oyarzún et al., 2015). In these papers, a metabolic enzyme is continuously controlled by a promoter and the enzyme converts a substrate into a final product. Two main feedback loop architectures have been considered: enzyme autoregulation or end-product feedback. Compared to a constitutive system with an identical mean product level, both enzyme autoregulation and end-product feedback could always reduce metabolite noise, with stronger feedback leading to stronger noise reduction (Borri et al., 2015). Additionally, feedback sensitivity was identified as a critical parameter to achieve large noise reduction for a wide range of promoters and feedback strengths (Borri et al., 2016, 2015; Oyarzún et al., 2015). This body of work provides an excellent theoretical foundation for the engineering of feedback systems for reduction of metabolite heterogeneity in the one-step pathways.
2.3.3. Controlling metabolite heterogeneity using population behavior control
Population behavior control is an emerging dynamic metabolic control strategy for controlling metabolite heterogeneity in engineered microbes (Lv et al., 2019). In this control paradigm, metabolite concentration is measured by a sensor which transmits this signal into a growth-modulating actuator (Fig. 1E). The actuator can provide a growth advantage to high-producing cells or steer low-producers towards self-destruction. In one example (Xiao et al., 2016), a FFA-based sensor-actuator was used to drive the expression of tetracycline efflux pumps in FFA-producing E. coli. Cells that stochastically produced low intracellular concentrations of FFA were unable to express sufficient efflux pumps, which caused cell death in the presence of tetracycline. Under the control of FAA sensor-actuator, the cell population displayed an increase mean FFA concentration and a 4.5-fold enhancement in FFA titer in a fermenter (Fig. 1E). This control strategy also proved useful to reduce phenotypic heterogeneity caused by genetic instability (Rugbjerg et al., 2018b). Here, a mevalonic acid (MVA)-based sensor-actuator was used to drive the expression of folP and glm, which are growth-critical genes for E. coli in a broad context. This control system eliminated cells that had low mevalonic acid production due to genetic heterogeneity, thus extending the productive lifetime of the bioprocess to 95 cell generations. Given the demonstrated promise of this control paradigm, we expect future developments in control theory for biological systems to address the benefits and limitations of population behavior control for dynamic metabolic engineering.
3. Sensors and actuation
Once a dynamic metabolic control topography has been selected, the individual parts and mechanisms need to be implemented. The essential parts of dynamic metabolic controls include sensors that detect metabolic or environmental signals, and actuators that change the output of the metabolic system in response to the sensor.
3.1. Sensing in metabolic control
Sensors are extremely useful in several metabolic engineering applications, which have been reviewed elsewhere (Liu et al., 2015a). This section will focus on sensors that have been demonstrated in dynamic metabolic engineering applications.
3.1.1. Sensing exogenous chemical signals
Common chemical inducers: Chemical inducers are traditionally used in open-loop pathways to induce the expression of metabolic enzymes but are also useful in two-stage switchable systems (Fig. 2A). Generally, cells are grown in a bioreactor to accumulate biomass, and a chemical inducer is added to halt growth as well as upregulate flux to a desired pathway (Burg et al., 2016; Soma et al., 2014). While being easy to use, chemical inducers can be expensive, must be added at a pre-determined time, and can be difficult to remove if switching the metabolic state back is desired.
 Fig. 2
Fig. 2Nutrient sensing: Many large scale bioprocesses choose to use various nutrients as the switching signal to reduce cost (Scalcinati et al., 2012). Glucose depletion, for example, is an attractive signal for two-stage fermentation. As glucose becomes completely consumed, cell growth ceases. Glucose-dependent promoters, such as those identified through genome-wide transcription datasets (Maury et al., 2018), have been used to induce metabolite production (Bothfeld et al., 2017). Other nutrient-based switching systems include systems induced by nitrogen-starvation (Sonderegger et al., 2005), phosphorus-starvation (Chubukov and Sauer, 2014), maltose (Liu et al., 2017), and ammonia (Xiao et al., 2017).
Quorum sensing (QS): QS is an attractive signal target for dynamic control since it naturally acts as a measure of cell population density which is linked to cell growth (Tan and Prather, 2017). When cell density passes a threshold, specific small molecules accumulate and can be used to induce a response to dynamically balance cell growth and target production while synchronizing cell activity in a population. Extensive work has been done to engineer QS for dynamic metabolic control. For example, in the gram-positive bacteria Bacillus subtilis, bifunctional and modular QS was engineered using the Phr60-Rap60-Spo0A QS system to improve production of menaquinone-7, a type of vitamin K (Cui et al., 2019). The quorum signal response can be tuned to different cell densities (He et al., 2017), thus enabling growth-to-production switching at different quorum for improved production of isopropanol (Soma and Hanai, 2015) (Fig. 2B), myo-inisitol (MI), and glucaric acid (Gupta et al., 2017).
3.1.2. Environmental signals
Temperature switches: Temperature-inducible systems have some benefits because the temperature of a vessel can be controlled externally, reducing the risk of contamination. Further, heat induction can be easily timed. Two heat-inducible systems extensively used in metabolic engineering are the λ phage promoters (i.e., PL and PR), both of which are regulated by the thermolabile cI857 repressor (Harder et al., 2018; Valdez-Cruz et al., 2010; Zhou et al., 2016, 2012) (Fig. 2C). Cold-inducible switches have also been developed via directed evolution (Zheng et al., 2019). Temperature induction also has limitations: (1) heat can alter protein folded structures and folding dynamics; (2) large or rapid change in temperature can trigger heat- or cold-shock responses and alter the global regulatory and metabolic networks; and (3) implementing temperature change in large fermenters uniformly can be slow due to heat transfer limitations.
Light switches: Optogenetic switches allow control strategies using visible light signals which is low cost and generally non-toxic. Light sensors are often derived from the photoreceptors domain of specialized proteins that undergo a conformational change in response to light (Milias-Argeitis et al., 2016). In an engineered Saccharomyces cerevisiae system, the light-responsive Phy/PIF module was adapted to allow red and far-red light pulses to switch on and off transcription of Gal4-responsive genes, enabling in silico feedback control (Milias-Argeitis et al., 2011) (Fig. 2D). Another engineered S. cerevisiae system used light to control fermentation with the Erythrobacter litoralis EL222 optogenetic transcription system, switching from a light-induced growth phase to a dark-induced production phase (Zhao et al., 2018). In an engineered E. coli system, the light sensor histidine kinase CcaS was used to respond to red and green light to control growth in an automated optogenetic feedback control system (Milias-Argeitis et al., 2016). A limitation of optogenetic switches is the problem of light-delivery since dense cell cultures scatter light significantly.
Other environmental switches: Several other environmental signals have also been used in dynamic metabolic controls. The Pgas promoter functions efficiently at pH 2.0 but becomes inactive above pH 5.0, thus was used to produce organic acid in Aspergillus niger, which further reduced pH of cell cultures (Yin et al., 2017) (Fig. 2E). The promoter of the cell wall glycoprotein CCW14 from S. cerevisiae was also engineered for a low-pH response to improve production of lactic acid (Rajkumar et al., 2016). Additionally, the oxygen-inducible nar promoter from the narGHJI operon in E. coli responds to anaerobic conditions and was used to produce 2,3-butanediol and 1,3-propanediol in E. coli when cell culture was switched to anaerobic conditions (Hwang et al., 2017).
3.1.3. Metabolite sensing
There is a high demand for endogenous metabolite sensors when building dynamic metabolic control systems. When engineered microbes can adjust their metabolism according to their intracellular metabolic status, each cell is spontaneously controlled even without synchronization at the population level.
Metabolite-responsive transcription factors (MRTFs): MRTFs are the most commonly used proteins for developing endogenous metabolite sensors. An MRTF usually binds to a specific metabolite and undergoes a conformational change that alters its DNA binding activity. Using the MRTF's cognate promoters or incorporating its operator sites into engineered promoters, transcription of metabolic genes can be controlled by the detected endogenous metabolite. Examples of MRTF-based sensors used in dynamic metabolic control include a FadR-based acyl-CoA sensor for FAEE production (Zhang et al., 2012) (Fig. 2F), FapR-based malonyl-CoA sensors for FFA and 3-hydroxypropionic acid (3-HP) production (Liu et al., 2015b; Xu et al., 2014; David et al., 2016), and a MexR-based pinene sensor for pinene production (Siu et al., 2018). The effector specificity of the MRTF can be altered through site-directed mutagenesis of the metabolite-binding pocket and screened by FACS as demonstrated in a HucR-based vanillin sensor for vanillin and ferulic acid production (Liang et al., 2020). Additionally, synthetic MRTFs can be created by fusing the DNA binding domain of a known TF with the ligand-binding domain from another enzyme (Chou and Keasling, 2013).
Stress-responsive promoters: For metabolites that cause cellular stresses, native promoters up- or down-regulated by the accumulation of the metabolite can be used as sensors. In E. coli, the accumulation of farnesyl pyrophosphate (FPP) from the engineered isoprenoid pathway is toxic (Martin et al., 2003). Stress-responsive promoters identified by genome-wide transcriptional analysis were used to control FPP-consuming enzymes, increasing amorphadiene production and improving growth (Dahl et al., 2013). Similarly, ergosterol-responsive promoters were used to dynamically regulate the expression of ERG9 in S. cerevisiae, leading to increased amorphadiene production (Yuan and Ching, 2015).
Other types of sensing: Besides MRTFs and stress-responsive promoters, aptamers present another mechanism by which a signal can be sensed (Liu et al., 2015a). While many aptamers exist and bind to a wide range of chemicals, the major hurdle is pairing metabolite-binding of an aptamer to an actuating mechanism while enabling tight control over gene expression and without disrupting protein function (Stevens and Carothers, 2015). Additionally, some metabolite-responsive signal peptides (also called leader peptide), such as TnaC that regulate the expression of downstream tnaAB expression in response to tryptophan, have been used as sensors for dynamic metabolic engineering (Fang et al., 2016) (Fig. 2G).
3.2. Actuation in metabolic control
In metabolic engineering, actuation usually involves controlling the abundance or activity of pathway enzymes in response to signals from the corresponding sensor. This section will discuss actuation strategies at different levels and highlight advantages and disadvantages with respect to the speed of flux response, and cellular economy utilization.
3.2.1. Transcriptional actuation
In addition to chemical inducers that are mostly used in two-stage fermentation (Burg et al., 2016), MRTFs can be directly used to repress or activate gene expression from their cognate promoters, enabling transcriptional actuation according to sensor signals. To obtain desirable actuation objectives, which are usually low leaky expression and high dynamic range, the cognate promoters controlled by MRTF often need to be engineered. This engineering effort can be guided by a phenomenological model that revealed the tunable parameters and constraints, such as the number and strength of repressor binding sites, in achieving the maximal dynamic range as well as low leakiness of sensor-actuators under various architectures (Mannan et al., 2017) (Fig. 3A). Another type of transcriptional control involves CRISPR interference (CRISPRi) or dCas9-mediated transcription repression. CRISPRi can be used to repress transcription of target genes upon binding to a sgRNAs, whose expression can be controlled by a sensor. For example, CRISPRi was integrated with glucosamine-6-phosphate (GlcN6P) sensors to enable sensing-actuation to control the metabolic flux of the nutraceutical N-acetylglucosamine (GlcNAc) pathway in B. subtilis (Wu et al., 2020) (Fig. 3B). Multiple orthogonal sgRNAs can be implemented into one strain to allow for multiple gene repression (Tan and Prather, 2017). When regulating multiple native genes is required, CRISPRi has advantages over MRTFs as there is no need for genome modification to change promoters of target genes. A drawback of CRISPRi is that the dCas9 expression must be carefully tuned to avoid toxicity caused by dCas9 overexpression.
 Fig. 3
Fig. 33.2.2. Post-transcriptional actuation
Gene expression can also be controlled post-transcriptionally using either antisense RNAs or RNA interference (RNAi). RNA repression has been used to regulate proB, glnA, and argB for their role in regulating intracellular ATP concentration (Chen et al., 2015). Hybridization between the antisense RNA and the RBS or open reading frame of the mRNA prevents translation or enhances mRNA degradation, ultimately decreasing gene expression. Antisense RNAs have been employed in E. coli for simultaneous regulation of multiple genes, autonomously redistributing carbon flux between native metabolism and an engineered muconic acid (MA) biosynthesis pathway (Yang et al., 2018) (Fig. 3C). In S. cerevisiae, the Argonaute (AGO1) and Dicer (DCR1) genes from yeast Saccharomyces castellii were used in conjunction with QS to degrade targeting mRNAs, enabling dynamic regulation of the shikimate pathway increasing para-hydroxybenzoic acid (PHBA) production (Williams et al., 2015) (Fig. 3D).
3.2.3. Post-translation actuation
Post-translational control of enzyme activity by a metabolite of interest can provide rapid control over the flux of a pathway and can be ideal for dynamic metabolic systems (Venayak et al., 2015). However, engineering allosteric enzymes for specific reactions is extremely challenging because novel allosteric interactions require high selectivity and robust output upon metabolite binding. Existing post-translation actuation mechanisms mostly focus on regulating enzyme degradation rate by adding degradation tags to target enzymes. For example, a TevP protease-based dynamic regulation circuit was constructed to create an ON and OFF switch that accumulates and degrades target proteins, respectively, depending on the degradation tags used. The protease was placed under growth- or stationary-phase promoters, allowing the system to respond to the growth stage and improving shikimate production (Gao et al., 2019) (Fig. 3E).
4. Applications of dynamic metabolic control
As theories and molecular biology tools are developed, an increasing number of dynamic metabolic control systems were developed for numerous pathways to benefit bioproduction (Table 1). In this section, we will summarize the pathways and metabolic nodes being dynamically regulated.
Table 1. Dynamic metabolic control in metabolic engineering.a