1. Introduction
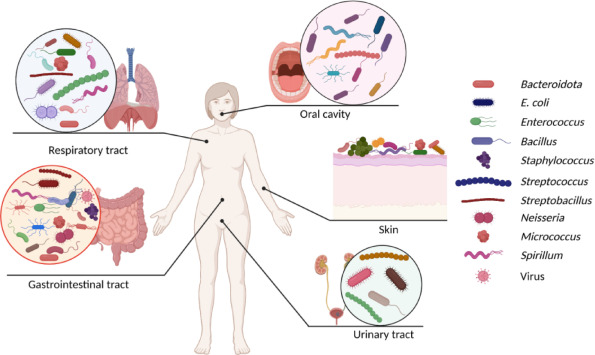
The human body houses 1013 to 1014 microbial cells, consisting of thousands of different species of bacteria, archaea, bacteriophages, viruses, and fungi [101]. These species form commensal, mutualistic, or pathogenic interactions throughout the entirety of the human host, such as on the skin and in the oral cavity, gastrointestinal tract, respiratory tract, and urogenital tract (Fig. 1). Most of these microorganisms are concentrated in the gut. The collection of these species harbors more genetic diversity than the human genome [108] and has been considered the “essential organ” of the human body [6]. Indeed, these species and their genomes, collectively named the microbiome, can regulate a variety of host processes, e.g., nutrient absorption, energy metabolism, drug metabolism, and immune responses. However, there are substantial variations in the densities of different species between individuals; many factors such as age, lifestyle, inherited genes, disease and geographic locations are all known to influence the human microbiome (Fig. 1) [38]. How exactly these variations affect human health and disease remains poorly understood but is currently an area of active and critical research. Nonetheless, changes in the microbiome are correlated with a wide array of illnesses, ranging from cancer, obesity, diabetes and mental health disorders to cardiovascular disease, inflammatory bowel disease, and infections [81]. Given the notable role of the human microbiome in maintaining and sustaining host health, microbiome engineering could be a powerful strategy for health improvement and disease treatment.
 Fig. 1. Diverse commensals, pathogens, and viruses are distributed at different sites of human body. The majority of the human-microbes interactions are found in the gut. Representative microorganisms in the microbiomes are indicated by the different shapes on the right, while different colors may be used in different microbiomes.
Fig. 1. Diverse commensals, pathogens, and viruses are distributed at different sites of human body. The majority of the human-microbes interactions are found in the gut. Representative microorganisms in the microbiomes are indicated by the different shapes on the right, while different colors may be used in different microbiomes.Microbiome engineering is a rapidly developing field that has received increasing interest for modulating human health as well as many other microbiome-associated sectors, e.g., ecosystem and agriculture sustainability [2,78]. In humans, the primary goal is to alter the function of the human microbiome by changing the composition of microbes or their genetic components (Fig. 1) [55]. As such, both native and foreign species, which may or may not be engineered, can be used for microbiome-based therapies. There are three major methods for microbiome manipulation: additive, subtractive, and modulatory approaches [76,115]. Additive approaches modulate the function of a microbial community by supplying individual strains or a defined microbial consortium, such as probiotic therapy [109] and fecal microbiotatransplantation (FMT) [57]. Subtractive approaches aim to eliminate specific pathogenic strains from the community. A representative example of the subtractive approach is the use of antibiotics to kill a broad spectrum of microbes. Modulatory strategies employ non-living agents or prebiotics to modify the activities of the endogenous microbiome [35].
Furthermore, certain organisms can be manipulated genetically to execute therapeutic functions that cannot be performed by endogenous strains [16,23,54]. The activities of these strains can be finely tuned and optimized in a predictable manner. For example, using synthetic biology methods, these species can be designed to detect signals of interest or to produce or deliver therapeutic agents in response to input signals [16,23,53,86]. Several human microbiome strains have been studied as model chassis in the past decades. The human probiotic E. coli Nissle 1917 (EcN) provides health benefits with a high safety profile [44] and is commercially available (Mutaflor®). Extensive research efforts have been devoted to EcN engineering to improve its efficacy or add new therapeutic activities [71,83,104]. For instance, antimicrobial activities of EcN have been enhanced by overproducing its native or non-native bacteriocins[37,83] or releasing quorum sensing molecules [28,46]. Furthermore, EcN can be engineered to produce and deliver anticancer agents in situ, such as agonists of stimulator of interferon genes (STING) [65], doxorubicin [120] and antibodies against immune checkpoint molecules [42]. Several other human commensals, such as Bifidobacterium, Bacteroides and lactic acid bacteria (LAB) Lactococcusand Lactobacillus, have also been engineered for therapeutic applications [9], although genetic tools for engineering these organisms are not as advanced as those for EcN. For example, members of the genus Bifidobacterium, which are obligate non-pathogenic anaerobes, have been engineered for cancer treatment through in situ activation of anticancer agents [99] and delivery or production of anticancer agents [66] and anti-inflammation cytokines [116].
Microbiome engineering frequently requires the creation of genetically engineered microbes, primarily bacteria. One essential step in genetic engineering is to deliver designed genetic materials into bacterial cellsefficiently. There are three major mechanisms of DNA transfer and uptake in bacteria: transformation, transduction, and conjugation [7]. Many genetic engineering techniques have been developed from these mechanisms to manipulate a variety of bacterial species and have become the cornerstone of modern genetics and molecular biology. However, no single technique is suitable to engineer all bacterial species [4]. In fact, except for a small number of model strains (e.g., EcN and Lactococcus lactis), many bacteria that can be cultured in the laboratory environment are not amenable to genetic engineering [51]. This is also the case when genetically engineering new primary isolates of undomesticated strains [11]. Two major hurdles in engineering undomesticated bacteria include the introduction of genes of interest and controllable and predictable gene expression [67]. In this regard, genetic intractability is a pervasive, fundamental challenge that prevents the basic and translational applications of microbes, including microbiome engineering. Here, we will discuss the fundamental aspects of transformation, transduction, and conjugation techniques in genetic engineering and present their applications in engineering human commensal bacterial species.
2. Transformation techniques for engineering microbiome bacterial species
Transformation is a process by which one organism uptakes naked DNA, potentially enhancing the fitness of host cells, supporting their DNA repair or supplying nutrition [17,74,91]. Acquired DNA can be incorporated into the host genome by homologous recombination (Fig. 2A), while some extrachromosomal DNA can replicate autonomously as plasmids, establishing a new episome[22,51]. The four basic steps of transformation include the development of competence, DNA binding to the cell surface, processing and uptake of naked DNA, and finally DNA chromosome integration (Fig. 2A). The detailed molecular mechanism of bacterial transformation has been reviewed elsewhere [18,22,50,107] and, thus, is not covered here. Genetic competence is a prerequisite to bacterial transformation. About 80 gram-positive and -negative bacterial species, such as Bacillus subtilis, Streptococcus pneumoniae, Neisseria gonorrhoeae, and Haemophilus influenzae, can undergo natural transformation to directly uptake naked DNA [17,51]. However, the vast majority of over 6600 validated, cultured type-strains of bacterial species are not naturally competent [32] and have to be prepared artificially for transformation. Unfortunately, each of these species, even each strain, generally requires an individually optimized protocol for the artificial transformation. Challenges to developing these individualized transformation approaches have led to the reliance on a handful of model strains.
 Fig. 2. A: General scheme of bacterial transformation. Permeabilized membrane of bacterial cells can be generated by both chemical and electroporation transformation. The transformed DNA can be integrated into the host chromosome or replicate as a plasmid. B: Schematic representation of engineering EcN (SYNB1618) to metabolize the high level of L-Phe-for treating phenylketonuria. Phe-transporter (PheP), phenylalanine ammonia lyase (PAL) and L-amino acid deaminase (LAAD) genes were integrated into the chromosome after electroporation.
Fig. 2. A: General scheme of bacterial transformation. Permeabilized membrane of bacterial cells can be generated by both chemical and electroporation transformation. The transformed DNA can be integrated into the host chromosome or replicate as a plasmid. B: Schematic representation of engineering EcN (SYNB1618) to metabolize the high level of L-Phe-for treating phenylketonuria. Phe-transporter (PheP), phenylalanine ammonia lyase (PAL) and L-amino acid deaminase (LAAD) genes were integrated into the chromosome after electroporation.To enhance the competence of bacterial cells, chemical transformation and electroporation methods have been developed and widely used (Fig. 2A). The first artificial transformation was performed with E. coli cells after CaCl2treatment in 1970 [72]. Subsequently, the transformation efficiency has gradually been improved to up to 109 colony forming unit (CFU)/μg DNA mainly by screening the use of different cations, optimizing cation concentrations, applying heat-shock and cold-shock, using organic chemicals, and tuning pre-competence cell growth conditions and cell growth stage [4]. The above factors can generate a smaller DNA packet for cell uptake by shielding the negative phosphate groups on the DNA backbone and lead to enhanced cell permeabilityand/or increased membrane flexibility. However, chemical transformation has been applied to engineer strains mainly from Proteobacteria (e.g., many model E. coli strains) and Euryarchaeota (e.g., Methanocaldococcus jannaschii) as the transformation efficiency is dramatically lowered by the presence of cell wall in many other bacterial species. To expand its applications, the cell wall can be removed to create spheroplasts or protoplasts that are then suitable for chemical transformation [8]. Many methods have been developed to partially or wholly remove the cell wall, including the use of divalent cations, polyethylene glycol, muralytic enzymes (e.g., lysozyme and mutanolysin) and EDTA. The success of the spheroplast or protoplast transformation has been demonstrated in the engineering of many gram-positive and -negative bacterial species, including various Streptomyces sp., Brevibacterium sp., Bacillus sp., Enterococcussp., Lactobacillus sp., Escherichia sp., Pyrococcus sp. and Methanococcus sp. [4,79].
Electroporation is the other commonly used transformation method that induces pore formation in diverse types of bacterial cells by applying electrical fields at around 5–10 kV/cm for 5–10 μs (Fig. 2A) [4]. The short but high-voltage pulses polarize the membrane components, thereby creating a voltage potential across the membrane [36]. After reaching a certain threshold level, the potential difference leads to the formation of local transient pores on the membrane at timescales of 10 ns, which allow the uptake of DNA. The pores are sealed within seconds, keeping cells alive. Electroporation usually demonstrates a higher transformation efficiency than chemical transformation (up to 1010 CFU/μg DNA) and can be used to engineer a significantly broader range of bacterial species, such as Bacillus sp., Bifidobacterium sp., Clostridium sp., Enterococci sp., Lactobacilli sp., Streptomyces sp., and Streptococci sp.. However, different species have distinct cell wall and membrane structures and different types and levels of DNA digestion enzymes, and thus optimization of electroporation parameters for each bacterial species is required to achieve high efficiency transformation [13,69].
Multiple microbiome bacterial species, particularly probiotics, have been engineered successfully by chemical transformation or electroporation methods. For example, EcN has been engineered to metabolize l-phenylalanine (Phe) to nontoxic metabolites by chromosomally expressing a high-affinity Phe-transporter (PheP) and two Phe-metabolic enzymes, phenylalanine ammonia lyase (PAL) and l-amino acid deaminase (LAAD) (Fig. 2B) [47]. The resultant strain SYNB1618 has been evaluated for the treatment of the rare disease phenylketonuria, which results in a high Phe-level in a patient's blood and brain. SYNB1618 recently demonstrated good safety and pharmacodynamicprofiles in phase I/IIa studies [89]. In addition to metabolism, EcN and other microbiome strains can be engineered to sense disease signals (e.g., nitrate and tetrathionate) and treat disease by expressing therapeutic proteins, host proteins/hormones, antigens and antibodies [42,53,65]. Both chemical transformation and electroporation methods have commonly been used to engineer these strains. For example, the SYNB1618 was created from EcN by electroporation (Fig. 2) [47]. Similarly, electroporation has been used to engineer the gram-negative bacterium Salmonella typhimurium to secret heterologous flagellin, which enhances cancer immunotherapy [121]. Furthermore, the gram-positive bacterium Lactococcus lactis has been engineered by electroporation to produce and secrete interleukin-10 for the treatment of murine colitis [105]. These successes demonstrate that the transformation can be a useful tool for engineering microbiome bacterial species. On the other hand, transformation has multiple limitations. For example, the transformation of naked DNA is often inefficient [106] as its success relies on the properties of the recipient strain and its amenability to electroporation or chemical transformation, both of which are largely unknown for isolated, undomesticated microbes abundant in the microbiome [41]. Furthermore, the plasmid systems used as the mobile genetic elements for preparing DNA for transformation are often specific to certain bacterial species, and sometimes individual strains, and have not been identified and developed for a large number of commensal strains [12]. In addition, transformed DNA may be degraded by the recipient's restriction-modification systems [111] and induced transformation cannot be used in situ for many commensals. Supported by an advanced understanding of the human microbiome, these limitations should be addressed to expand the use of transformation in microbiome engineering.
3. Phage-based techniques for engineering microbiome bacterial species
Viruses that infect bacteria, named bacteriophages or phages, are the most abundant organisms on Earth and can be found at 10 times the abundance of bacteria [77]. They are present in almost all microbial communities, including the human microbiome (Fig. 1) [14,102]. The vast majority of bacteria can be infected by these obligate intracellular parasites [21]. A phage contains either a DNA or RNA genome, surrounded by a protein coat (Fig. 3). When infecting bacteria, the phage injects its genetic material into the bacterial cell, which is then recombined into the bacterial chromosome or replicated like a plasmid (Fig. 3). For lytic or virulent phages, the phage genome is immediately replicated and new phage particles are assembled to lyse and kill the host, releasing 100 to 3000 progeny phages [26]. In contrast, temperate phages can adapt lysogenic and lytic cycles (Fig. 3), leading to their temporal stay in bacterial cells. In the lysogenic cycle, the phage genome is integrated into the host chromosome and is vertically inherited when the host cell divides. Temperate phages can be induced by some conditions (e.g., nutritional availability, antibiotic treatment, and oxidative stress) to enter the lytic cycle, in which they kill the host cells and release viral particles (Fig. 3) [29]. When entering the bacterial cells, the phage genome provides new genetic material to the host, which can affect host fitness (e.g., protecting the host from lytic infections) and enrich virulence traits [33], thereby modulating bacterial populations.
 Fig. 3. Schematic of phage-mediated DNA transduction. After lysogenic induction, pieces of host chromosomal DNA can be packaged and passed to new bacterial cells through specialized and generalized transduction.
Fig. 3. Schematic of phage-mediated DNA transduction. After lysogenic induction, pieces of host chromosomal DNA can be packaged and passed to new bacterial cells through specialized and generalized transduction.When propagating, temperate phages can pick up host genes and carry them to a new bacterial host (Fig. 3). The bacterial DNA can be maintained in a new host cell through chromosomal integration or replication like a plasmid. This process is known as transduction and, despite occurring at a low frequency, is an important mechanism of horizontal gene transfer among bacteria [21]. Phage-mediated exchange of bacterial DNA can undergo specialized and generalized transduction (Fig. 3). In specialized transduction, transducing phage particles contain a defective, hybrid phage genome that loses part of the original phage genome but carries bacterial DNA adjacent to phage attachment sites (attB) [21]. Lambda phage is a typical specialized transducer [52], which is made at a frequency of 1 in 104 virions [59]. In generalized transduction, phages may transfer any piece of the bacterial host genome in place of their own [87]. The DNA packaged into transducing particles for generalized transduction depends on the presence of the homologs to the pac site sequence scattered throughout the bacterial chromosome and plasmids, which are recognized by the phage small terminase (TerS) [87]. Multiple phages can carry out generalized transduction, such as P1 and P22. The frequency of generalized transduction is still low as only 1% to 6% of virions contain host DNA, but is significantly higher than that of specialized transduction [80]. Nonetheless, both specialized and generalized transduction are important routes of bacterial DNA exchange in almost all ecological niches. For example, a recent study of a murine gut metagenome suggests that close to 9% of bacterial contigs carry transduction signals and a significant number may be different from any known mode of transduction [59], highlighting the incomplete understanding of phage transduction. Such a paucity can negatively influence the use of phages as tools to manipulate the microbiome. Indeed, phage-mediated horizontal gene transfer can be a key co-evolutionary driving force for the gut colonization of introduced bacteria and for the promotion of genetic diversity of commensal bacteria in the gut [34].
As an important component of the human microbiome, both virulent and temperate phages can regulate bacterial abundance, diversity, functions, and evolution. Importantly, they can be used to manipulate the human commensal microbes [56]. For example, a Phase I/II clinical trial has confirmed the efficacy of biophage PA in treating chronic otitis caused by drug-resistant Pseudomonas aeruginosa [114]. Furthermore, in a recent case study, the combined use of conventional antibiotics and two lytic phages PA3 and PA18 successfully alleviated the infection caused by carbapenem-resistant P. aeruginosa in a patient with empyema-associated broncho-pleural fistula [19]. Similarly, multiple phage-based therapies have successfully treated infections in humans and animals caused by pathogens such as Acinetobacter baumnnii [100], Klebsiella pneumoniae [40], and Clostridium perfringens [75]. In addition to targeting a single pathogen, phages can be used to regulate a variety of bacterial strains in the same microbiome for improving health. For example, viral communities from lean mice successfully reduced the symptoms of type 2 diabetes and obesity in recipient obese mice [90], while sterile-filtered stool samples of healthy donors, which carry diverse phages, showed positive outcomes in treating Clostridium difficile infection in a small number of patients [82]. In these applications, admitted phages often infect only a single bacterial species or multiple strains of the same species, demonstrating high specificity [77]. Specific clearing of a given bacterial strain/species in the microbiome is particularly useful in precise disease treatment when causative bacteria are known. Unfortunately, this is not the case in most dysbiosis-associated disorders, challenging the use of phages for rational microbiome engineering [112].
Another challenge of phage-based microbiome engineering is the limited number of well-characterized phages. Among about 1031 phages on Earth, only several thousand have been isolated and they infect only a small number of bacterial phyla [117]. Given their high host specificity, these known phages offer limited options for the precise engineering of the human microbiome. On the other hand, phages also feature high evolvability that lends itself to opportunities for the rational engineering of known phages for new applications [58,61]. For example, the host range of phages can be altered and extended by modifying receptor-binding proteins on their tail/tail fibers that interact with the cognate bacterial surface receptors. The modifications can be performed by random mutagenesis [70], rational engineering [118], and/or swapping of different phage tail/tail fibers [3,62,119]. This synthetic biology strategy has demonstrated success in engineering the host range of multiple gram-negative and -positive bacteriophages, e.g., P1, T2, T4, T7, and PSA. Another strategy in phage engineering is to equip phages with needed functions for manipulating the microbiome. For example, the Turnbaugh group recently engineered the filamentous bacteriophage M13 to deliver a programmable exogenous CRISPR-Cas9 system to E. coli in the mouse gut, leading to strain-specific depletion [63]. These examples showcase the promise of phage engineering for microbiome manipulation. More details and synthetic biology strategies for phage engineering have been reviewed recently elsewhere [20,45,58,88] and are thus not covered here.
4. Conjugation techniques for engineering microbiome bacterial species
When engineering microbes, especially undomesticated microbes, one of the primary causes of genetic intractability is thought to be the destruction of the foreign DNA of interest by the recipient microbe's defense systems, e.g., restriction-modification, CRISPR-Cas, prokaryotic Argonaute, and more [94,111]. Thus, the initial steps of introducing foreign DNA and preventing its destruction within the recipient are vital to the successful engineering of a new microbe.
Conjugation is a widespread method of horizontal gene transfer between prokaryotes [68] and overcomes many host-cell barriers to foreign DNA introduction, making it a useful method for microbial engineering (Fig. 4). It is a cell-to-cell contact-dependent, single-event transfer of DNA fragments that can be large and nonhomologous to the recipient's genome [10,41]. Conjugative systems are found in both gram-positive and -negative bacteria and are made up of Type 4 Secretion System (T4SS) machinery. Conjugative machinery is encoded either by self-transmissible conjugative plasmids (CPs) or integrated conjugative elements (ICEs) (Fig. 4) [5,11,39,41,60]. ICEs, also known as transposons, are the most abundant self-transmissible elements [60]. Similar to temperate phages, they are maintained chromosomally until their gene expression and excision are induced [27,39]. Upon excision, the ICE undergoes rolling-circle replication and can be transferred to a recipient as a plasmid. In gram-negative and most gram-positive bacteria, the transmissible plasmid is nicked and then threaded through the T4SS as a protein-ssDNA complex into the recipient [10,39,60]. A major benefit of the introduction of the plasmid as ssDNA is the synthesis of the complementary strand by the recipient with host-specific modifications (Fig. 4) [97]. As a result, the recipient does not recognize the plasmid as foreign, allowing it to bypass defense systems. Therefore, conjugation is an attractive option for engineering undomesticated microbes as it provides the direct introduction of DNA into the recipient's cytoplasm [111], bypasses recipient defense mechanisms [97], and can be used for in situcommensal microbe engineering [11,24,94,96]. Indeed, conjugation has broader applications in current microbiome engineering, compared to transformation and phage-based engineering.
 Fig. 4. Schematic of conjugation. The donor cell contains the coding for conjugative machinery integrated into the genome or on a replicative plasmid. If integrated into the genome, the ICE is first excised into its plasmid form. The donor cell then establishes cell-to-cell contact with a recipient via a pilus (gram-negative bacteria) or adhesins (gram-positive bacteria) and transfers the DNA. The recipient cell then contains the new DNA, either integrated into the genome or as a replicative plasmid.
Fig. 4. Schematic of conjugation. The donor cell contains the coding for conjugative machinery integrated into the genome or on a replicative plasmid. If integrated into the genome, the ICE is first excised into its plasmid form. The donor cell then establishes cell-to-cell contact with a recipient via a pilus (gram-negative bacteria) or adhesins (gram-positive bacteria) and transfers the DNA. The recipient cell then contains the new DNA, either integrated into the genome or as a replicative plasmid.Multiple different conjugative machineries have been developed and implemented within the laboratory for engineering microbes. When choosing a conjugation system, one important factor is the host range [24,39,60,93,106]. Traditionally, conjugative and mobilizable plasmids were classified as either having broad or narrow host ranges, indicating the number of hosts that the plasmids could be introduced into and maintained in [92]. A recent analysis of conjugative plasmids indicates that there is a larger gradation of the number of host microbes in which these plasmids can be found. Furthermore, conjugative transfer systems often decrease in effectivity with greater phylogenetic distance unless they are more promiscuous [11,73,92]. Thus, identifying the range of microbes that one is targeting is important in selecting a conjugative system. Finally, gram-negative and -positive conjugative T4SSs differ in structure and requirements for function. Gram-positive bacteria typically contain a simpler T4SS, while gram-negative bacteria have a more complex T4SS that can span two membranes [39,60]. When establishing contact between the donor and recipient cells, gram-negative bacteria form pili to pull the two cells into close contact while gram-positive bacteria use adhesins that are surface-exposed binding proteins (Fig. 4) [10,39]. In addition, in most bacterial cells, there are layers of peptidoglycan outside the plasma membrane, which consist of sugars and short peptides. The thickness of peptidoglycan in the donor and possibly the recipient is also an important consideration as the peptidoglycan hydrolasesfound in gram-negative conjugative systems only have a single catalytic domainand are not required for conjugation, while the peptidoglycan hydrolases in gram-positive conjugative systems have two or more catalytic domains and are required for conjugation [10,27,39,60]. To achieve the broadest host range, a conjugative system may need to span two cellular membranes and target gram-negative bacteria as minimized T4SSs have been found to work in a variety of undomesticated gram-positive bacteria but not gram-negative bacteria [11].
Another consideration when choosing a conjugative system is the maintenance method of the delivered gene construct (Fig. 4). While plasmids can be useful for short-term or transient gene maintenance, integration of the target gene(s) into the genome lends greater genetic stability [110]. Conjugative systems can easily mobilize non-conjugative plasmids that contain the appropriate origin of transfer (oriT) [60] but integration of the target genes often requires additional steps. Incorporating the gene(s) of interest directly into the ICEs allows for concurrent integration into the recipient genome. Some ICEs target specific integration sites along the genome, such as ICEBs1 targeting the leucine tRNA gene [27], while other transposons incorporate randomly, such as Himar or Tn5 [85,94]. Random integration can be useful for nonspecifically targeting a wide range of hosts, but it has its shortcomings as integration location contributes to heterogeneous expression [85]. In this regard, the addition of a catalytically dead CRISPR-Cas protein (dCas) can be used to guide transposons to specific sites for integration [96]. Similarly, the use of integrases, recombinases, or homologous recombination can be explored as a secondary step of gene introduction [110,113] and can also be guided using dCas [110]. A disadvantage of specific integration is the required knowledge of the targeted bacteria's genome. Nonetheless, highly conserved, duplicated genes such as 16S rRNA genes can provide an ideal site for integration if a specific bacterial species is being targeted [49].
As the fields of synthetic biology and microbiome studies advance and intertwine, researchers are looking outside of domesticated, model microbes to undomesticated microbes as a means to elucidate the impacts and mechanisms of microbiome functions [1]. Conjugation has provided an effective and generally applicable method for engineering uncharacterized microbes. Studies have demonstrated that conjugation can be used to introduce genes into multiple bacterial isolates, demonstrating the broad host range of this method [11,49,113]. For example, two recent studies developed CRISPR/Cas-based genome editing approaches to engineer multiple human gut commensal Bacteroides species [30,122]. To achieve high editing efficiency, these authors evaluated multiple Cas nucleases (e.g., Cas9, FnCas12a, SpRY, and FnCpf1), constitutive and inducible promoters, ribosome binding sites, and other genetic elements. All constructs were then introduced into Bacteroides species through conjugation. Furthermore, conjugation has been used to engineer undomesticated bacteria in complex communities, eliminating the need for prior strain isolation [10,24,96,110]. In fact, conjugative methods have been applied to screen and enrich genetically tractable undomesticated bacteria in these complex communities (e.g. stool samples, soil samples) [24,96] and even in situ in the murine gut microbiome [94].
Though conjugation is proving to be a powerful tool for microbial and microbiome engineering, there are still limitations that must be considered. The recipient host range of each conjugative system is a limiting factor, with even the most promiscuous conjugative plasmids in class IncP1 being limited mainly to the order Burkholderiales [92]. For widespread targeting of a microbiome, the use of multiple conjugative systems may be necessary to achieve the desired spread. However, conjugative plasmids are organized into incompatibility classes (Inc classes) in which a recipient bacterium cannot receive a new plasmid if it already harbors a plasmid of the same incompatibility class [15,60,97]. When considering gene construct maintenance, the origin of replication must be appropriate to the host species if using a plasmid; otherwise, integration can be used instead, either randomly or targeted, which comes with the limitations discussed previously. Finally, successful expression of the gene of interest can be host-specific [30,122], which is why genes are often codon-optimized and used with host-specific regulatory factors when introduced into domesticated laboratory bacteria such as E. coli MG1655. Re-designing a gene using one established codon-optimization table or with particular regulatory elements can limit its ability to be expressed and detected in other microbes, even if it is successfully introduced. Alternatively, recent work has shown that synthetic, hybrid regulatory elements and generalized codon-optimization could be the path forward for the design of “universal” gene constructs that work across kingdoms [85].
5. Conclusions and perspectives
Synthetic biology applications in microbiome research is expected to have a wide range of basic and translational applications. Genetic manipulation provides direct, top-down approaches to probe the functions of individual microbiome strains or microbiome communities. Widely used in nature, transformation, transduction, and conjugation are three major methods that allow for successful microbial or microbiome genetic manipulation. Many studies have reported the protocols and new development of these methods in engineering many different bacterial species. However, each of these methods has its advantages and limitations, which are summarized in Table 1. For example, transformation has high efficiency but its host range is relatively narrow [4], while transduction has a high host specificity but phages specifically infecting a given bacterium are often unidentified [117]. Currently, microbiome engineering requires the testing and optimization of all of these methods to promote success. For example, a recent work developed a genetic manipulation pipeline for the engineering of nonmodel commensals [49]. The combined use of conjugation and electroporation methods led to the successful engineering of 88 (mostly undomesticated) out of 200 gut bacterial isolates from over 140 species in five phyla. Of note, these gene transfer methods were applicable to engineer 38 nonmodel gut Clostridia isolates that are abundant in human guts but lack effective genetic manipulation tools.
Table 1. Summary of major advantages and limitations of transformation, transduction, and conjugation in DNA introduction for the engineering of commensal microbes.
| Method | Major advantages | Major limitations |
|---|---|---|
| Transformation | Tools and protocols have been well developed for multiple model strains. |
The majority of bacterial species are not naturally competent; Transformation efficiency into undomesticated bacteria is often low; Species-specific genetic systems are often required. |
| Transduction |
High specificity can be used to develop precise manipulation; The high evolvability of phages offers opportunities for extending the host range and functions. |
The number of well-characterized phages is limited; Species-specific targeting restricts the wide manipulation of the microbiome for treating most dysbiosis-associated disorders. |
| Conjugation |
It has broad applications in engineering single commensal isolates and specific strains in complex communities; Transferred DNA can be large and nonhomologous to the recipient's genome; ssDNA introduction bypasses recipients' defense systems. |
The recipient host range of a conjugative system is limited; Incompatibility class plasmids prevent the use of multiple conjugative systems; Expression of the gene of interest can be host-specific; Genome integration can be random or requires the knowledge of the host genome. |