1. Introduction
Fluorescence is a luminescence process in which atoms and molecules absorb a specific lower wavelength light and after a brief gap of fluorescence lifetime emit a longer wavelength light. Over the past many years in the service of biology, fluorescence microscopy has emerged as the prime pillar of microscopy due to its inherent selectivity to expose only objects of interest against black background [1]. To investigate myriad of cellular processes, fluorescence imaging has been realistic for the visualization of molecules and whole organisms. It started with the attachment of organic dyes to proteins through antibodies and later sought the genetic tagging of target proteins with fluorescent proteins. However, the linking of antibodies has to be supplemented with the fixation and permeabilization of cells [2]. With the efficacy of fluorophores as direct recognition agents for various cellular molecules like nucleic acids, ions and other cell organelles, the technique of immuno-fluorescence became less valuable. Additionally, with the development of FRETbased sensors, noninvasive behavior and live cell imaging in both prokaryotic and eukaryotic cells proved to be a novelty for researchers exploring fluorescent probes for real time cellular imaging. Originally, FRET stands for Forster Resonance Energy Transfer in the honour of Theodor Forster, physical chemist, who first discovered and understood it. The term “fluorescence resonance energy transfer” is often used, when both the chromophores are fluorescent. The latter enjoys common usage in scientific literature and has been in practice as such commonly.
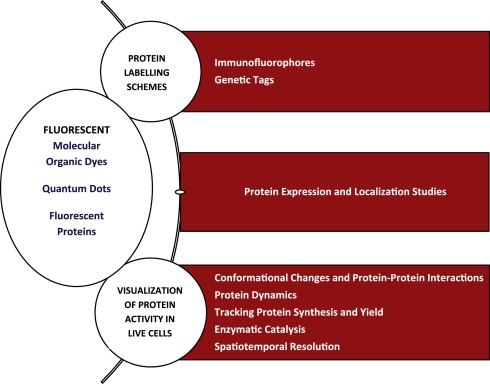
However, It has been established that compared to other fluorophores, amplified brightness and photo-stability are the two critical parameters of semiconductor nanocrystals that makes them unique for fluorescent microscopic imaging (Fig. 1), although their targeting potential still endures many hindrances [3]. In recent years, using newly engineered photo-controllable fluorescent proteins; numerous super-resolution microscopic techniques became ready to use in order to get better visualization of objects having dimensions smaller than 500 nm and 200 nm in axial and lateral directions, respectively. In current times, researchers are now powerfully aided with more precisely optimized FPs. There are three groups of photo-controllable FPs- photo-convertible FPs (PCFPs), photoactivatable FPs (PAFPs), and reversibly photo-switchable FPs (rsFPs). PAFPs are activated from non-fluorescent (dark) to fluorescent state, whereas, rsFPs are reversibly photo-switched between inactive and active states, and PCFPs are made to convert from original color to another. The super-resolution microscopic imaging using these FPs is always carried out by controllably turning them on and off [4].
 Fig. 1. Outline of fluorescent probes, their types, applications and different methods to study protein dynamics and localization within a cell or whole organism.
Fig. 1. Outline of fluorescent probes, their types, applications and different methods to study protein dynamics and localization within a cell or whole organism.Moreover, proteases have emerged favorable enzymes for synthesizing their inhibitory compounds as potential drugs for some major human diseases such as cancer, AIDS and other neurodegenerative infections. Because of the latest coherent advancements between genomic, proteomic and biophysical techniques, peptide substrates for several proteases have been ascertained, that paved a way to trace and analyze the functioning of their equivalent proteases through a simplistic and speedy mode. By combining such peptide substrates with appropriate FPs, researchers develop a chimeric protein and then sequentially analyze many protease inhibitors through FRET disruption. Present review will focus on latest advances in major fluorophores and diverse fluorescence strategies that are in current use of fluorescence microscopy to visualize protein dynamics involved in their location and function within a cell.
2. Variety of fluorescent probes
Varieties of fluorophores that are beyond the scope of this review are now available for evaluating the protein activity. Regarding fluorescent probes, two critical considerations must be acknowledged first the fact that spectral properties of fluorescent probes determine the basic settings of time resolution and wavelength of instruments, and secondly certain fluorescent probes are used to monitor specific activities. For example, fluorophores that are sensitive to pH can only be explored to measure pH, and rotational diffusion can be tracked by only those probes having non-zero anisotropies. For histological studies, probes possessing long excitation and emission wavelengths are utilized to display auto-fluorescence at short excitation wavelengths. We are elaborating here some basic fluorophores which are being exploited through various bioengineering approaches to visualize the sea of protein dynamics, so as to come up with innovative nanoscale toolkits for monitoring molecular interactions operating within a living cell in real time.
2.1. Molecular organic dyes
In the synthesis of organic fluorophores, many molecular strategies have been adapted. For this purpose, addition of electrophiles or other charged substituents like sulfonates, conjugation of double bonds, extra ring rigidification by locking rotatable rings into parent ring structures, etc. are some of the most important assignments. Though the dyes made by such strategies are now available commercially (Haugland, 2005), however, such dyes are handicapped by being non-specific to all proteins. Therefore, their use in permeabilized and fixed cells has to be supplemented with antibodies (Fig. 2A). These organic fluorophores are usually of low magnitude (<1 kD) and their important etiquettes like reduction in self-quenching, good brightness and appropriate wavelength have made them industrially optimized.
 Fig. 2. Application of different types of fluorophores, exploited for labeling and detection of proteins. (A) A 3T3 cell from an adherent culture, immunolabelled with Alexa Fluor 488. (B) Quantum Dot with CdSe core and ZnS shell. (C) A QD conjugated with many antibodies, preventing its mobility. (D) A ligand sensing domain, inserted between CFP and YFP.
Fig. 2. Application of different types of fluorophores, exploited for labeling and detection of proteins. (A) A 3T3 cell from an adherent culture, immunolabelled with Alexa Fluor 488. (B) Quantum Dot with CdSe core and ZnS shell. (C) A QD conjugated with many antibodies, preventing its mobility. (D) A ligand sensing domain, inserted between CFP and YFP.For a successful delivery of fluorophore into a cell, broadly two types of cellular environments- intracellular and extracellular are available. The major confrontations to overcome lies in optimized intracellular delivery of fluorophores, the perfect labeling of target biomolecule and negligible cytotoxicity. Due to smaller size of organic fluorophores there is least spatial hindrance to impede with functioning of target biomolecule. Therefore, this opportunity has been explored by attaching many fluorescent probes with a single target biomolecule in order to gather maximum fluorescence signal. Because of high label densities the strong electrostatic repulsion between neighboring molecules, the dye structural conformation and hydrophilicityaltogether can cause fluorescence quenching [[5], [6], [7]] and may also affect functioning of the biomolecule [8].
For the selective detection of enzymatic and non-enzymatic proteins, fluorescent chemical turn-on probes are synthesized by fusing an environment sensitive fluorophore with a protein-specific small molecule [9]. In this scenario, localized deposition of proteins in proteostasis has been elaborately exploited to develop sensors for understanding the mechanism of protein deposition in neurodegenerative disease progression [10]. The process of proteostatis involves biogenesis, traffic and protein degradation within and outside a cell, involving different integrated biological pathways. The basis of such a designed fluorescent probe is that most of the ligand binding sites in proteins are hydrophobic, that constitute the prime thermodynamic driving force for the binding of small molecule ligands to their respective proteins. This approach has been exploited to develop a precious turn on folding sensor for effective live cell monitoring of proteostatis [11]. This small fluorogenic molecule became fluorescent when it binds and reacts with folded and functional retroaldolase enzyme. Recently, in a similar approach, dual signal fluorescence-enhanced sensor, based on Cu2+ mediated fluorescence switchable strategy has been designed to detect Cysteine (Cys) in a simple and fast way. It was observed that two fluorescence emissions of ultrathin films (calcein@NFR/LDHs UTFs) are effectively quenched by Cu2+ (off state), and then reversibly recovered by Cys (on state), leading to the specific coordination of Cys and Cu2+ [12]. Similarly, a novel, simple and rapid fluorescent probe based on excited-state intramolecular proton transfer (ESIPT) was the latest addition to such Turn On sensors, allowing easy way detection and quantification of biothiols in living cells [13].
For site specific protein labeling in live cells, Roger Y. Tsien and colleagues in 1998 pursued the use of biarsenical reagents. In real sense, it is the high affinity interaction of arsenic for thiols that forms the basis of biarsenical labeling technology. The fluorescent derivative FlAsH contains two arsenic atoms at a set distance from each other. Similarly, ReAsH is modified to contain resorufin. With the aid of fluorescence microscopy, FlAsH and ReAsH technology has been exploited to bind to tetracysteine (TC) sequences. The common TC sequence employed for this purpose is the six aminoacid sequence Cys-Cys-Pro-Gly-Cys-Cys. When bound to ethane dithiol (EDT), both FlAsH and ReAsH are non-fluorescent. Thus, upon binding with recombinant proteins containing such a tetracysteine motif, both these biarsenical labeling reagents- FlAsH-EDT and ReAsH-EDT turn to become highly fluorescent with green or red color respectively, with subsequent displacement of EDT.
Another most important bacterial enzymatic protein Haloalkane dehalogenase a hydrolase is modified and designed to covalently bind to a synthetic ligand with subsequent fusion to the protein of interest. Therefore, this enzyme has been efficiently exploited for visualizing subcellular localization of protein of interest, to capture the binding partners of a protein or for protein immobilization [2]. The function of such a single protein tag is altered by attaching different chemical moieties of synthetic ligands called HaloTag ligands, through a chloroalkane linker, attached to useful molecules such as fluorescent dyes, affinity switches or solid surfaces [14]. The Halotag is a modified haloalkane dehalogenase designed to covalently bind to synthetic ligands (Halotag ligands). It needs to be understood that, covalent bondformation between FP tag and the chloroalkane linker is extremely specific, occurs swiftly under physiological settings, and is essentially irreversible. However, the choice of ligand is done in accordance with the type of experiments to be performed.
Because of the heavy load of metal pollutants, researchers have come up with some important electrochemical sensors to detect different metals in a specific, sensitive and selective way. For example, to detect attomolar (aM) concentrations of Hg2+ a precious electrochemical sensor was developed [15]. For detection of this target metal, three single-stranded DNA probes were rationally designed. These probes were developed due to combined thymine-Hg2+-thymine (T-Hg2+-T) based coordination chemistry principles. In a similar approach, a sensitive electrochemical lead ion sensor was developed for lead (Pb2+) ion detection. It was observed that, it was due to Pb2+-induced G-rich DNA conformation that upon Pb2+ addition, DNA duplex got unwound and formed a stabilized G-quadruplex (G4) [16]. Another approach for detecting Pb2+, was based on an electrochemical DNA sensing strategy through modification of a glassy carbon electrode, with ordered mesoporous carbon nitride, gold nanoparticles and methylene blue [17].
2.2. Quantum dots
Quantum dots (QDs) contain some hundred to thousand atoms within a size of nanometer scale, distinctively made of an element of silicon or germanium or composed of a core of CdSe or CdTe and a ZnS shell (Fig. 2B). Such petite structures vary in their color. The sharp fluorescence of such nanocrystals at discrete wavelengths depends on their size. Compared to other fluorophores, QDs possess 10–100 times higher extinction coefficients and better quantum yields [18]. Here, a lone excitation wavelength readily excites the QDs of numerous emission spectra. QDs are preferably developed with coatings for biological investigations to facilitate their solubility in aqueous medium, help in conjugation with antibodies and prevents quenching by water [[19], [20]]. However, QD conjugated biomolecules lack efficient migration potential through intact cellular membranes due to their larger size, and thus, their expenditure for endocytosed proteins or extracellular apartments, permeabilized cells is restricted. Remarkable about QDs is their tolerance to repetitive imaging of solitary molecules, by virtue of their photo-stability [21]. The portable electron-density and size permits analogous electron microscopyto be used for visualization of different objects of interest. However, a major issue with QDs is their toxicity in biological applications, and construction of conjugated polymer dots has helped a lot in this way.
Concerning the use of QDs to label target biomolecules, there is no strict protocol. However to achieve an optimized labeling, QDs are first made to become water dispersible and are then attached to target biomolecules. Many interactions are exploited to achieve QD fluorophore labeling with target biomolecules. Some major linking methods include-covalent linkages, biotin-avidin interactions and poly-histidine tags. Compared to small organic fluorophores, many biomolecules are attached to a single QD [22] which leads to problematic orientation (Fig. 2C). On the other hand, because of the better cell and organelle permeability of aryl fluorosulphates, they have been employed for the development of protein-selective covalent probes. In this scenario, an environment sensitive fluorogenic probe, 1,3,4-oxadiazole has been designed to bind selectively to transthyretin (TTR). Addition of this fluorogenic probe to HEK293T cells allowed efficient binding and imaging of cellular organelles like mitochondria and endoplasmic reticulum, making it a new fluorescent tool for living cells [23]. Upon application of this fluorogenic probe in Caenorhabditis elegans, it successfully detected TTR in six macrophage like cells.
2.3. Fluorescent proteins
In real sense, the breakthrough of green fluorescent protein (GFP) from Jelly fish (Aquorea victoria) and then subsequent engineering of various other novel FPs from diverse organisms armed us with fluorophores, possessing extraordinary uniqueness for live cell imaging. Single GFP based sensors or chimeric FRETbased nanosensors generate visible fluorescence for microscopic imagery. FRET involves the transfer of non-radiative energy from donor fluorophore to acceptor fluorophore, in the presence of a ligand. Such novel approach has been utilized to develop chimeric proteins that serve as nanosensors. These FRET based nanosensors consist of cyan fluorescent protein (CFP), a ligand binding domain and a yellow fluorescent protein (YFP) (Fig. 2D), with CFP and YFP as the two mutant forms of GFP. However, after GFP, large spectrums of FPs with varied colors were discovered from marine coelenterates. The increase in brightness and folding efficiencies, accompanied with a decrease in oligomerization is achieved by generation of affinity mutants. This leads to the diversification of spectral range of FPs, from one color to another, improving the overall protein monitoring system of the resultant biosensor. Mutagenesisincreases both photo-stability and photo-switchability of FPs [[24], [25]]. It is exactly the reversibility and irreversibility in photo-switching that makes FRET based biosensors useful to track protein trafficking. Quenching enforced by acidic pH is the major snag in the bio-sensing mechanism of FPs. However, recent developments have now better engineered the sensitivity of biosensors vis a vis ions, pH and redox potentials [[26], [27]].
Prior to FRET based biosensors, phyco-bili proteins and cyanobacterial photosynthetic antenna pigments were used as prime tags for fluorescence. It is due to their bigger size that problems arise in their diffusion which renders them limited only for surface labeling purposes [28]. Therefore, such FPs are routinely utilized in conjugation with antibodies for enzyme-linked immunosorbent assays and flow cytometric measurements. For the FPs to offer their best character as fluorophores, some vital features include- varying spectral properties, maturation efficiency, photo stability, efficient brightness, and fidelity in fusions, monomeric character and their potential efficiency as FRET donors or acceptors. Furthermore, critical mutations have been exploited to generate different FP variants with enhanced spectral properties- mVenus [29] and mKO2 [30] are the two important examples of bright monomeric variants. To address spectral and structural snags of FPs, recently a family of GFP proteins in the cephalochordate Branchiostoma floridae, commonly known as amphioxus was discovered and named after it as bfloGFP [31]. So far, this animal is credited to be the only species representing the largest source of 16 FPs. A total of six clades of amphioxus possess these 16 GFPs, all emitting the green fluorescent light, and each clade owns discrete absorption spectra, extinction coefficients and fluorescence intensities. Utilizing the x-ray crystallography derived three dimensional (3D) structures, biochemical and spectral characteristics of two FPs- a bright FP (bfloGFPa1) and a weak FP (bfloGFPc1), this group [31] deliberated about the role of structural differences in FPs vis a vis chromophore environments that modulate into their photonic properties. A latest approach [32] has been carried out to improve the brightness and photostability of green and red FPs for enhanced live cell imaging in FRET. The group reported an improved photostability of mClover3 a derivative of GFP and mRuby3 a derivative of Red Fluorescent Protein (RFP) by an extent of 60% and 200% respectively over the previous generation of fluorophores. Also mRuby3 was recorded to be 35% brighter than its previously engineered version- mRuby2. Till the present period so far, out of all Jelly fish GFP and coral RFP derivatives, expressed in various mammalian cell lines, mClover3 and mRuby3 offers the highest fluorescence signals.
3. Protein labeling schemes
In order to obtain the visualization of the protein of interest, two tagging schemes are generally followed- fluorescently labeled antibodies, in which antibodies are made to specifically bind with the protein of interest and the intrinsic fluorescent signal, where FP is genetically linked with the protein of interest. Though, in comparison to active antibodies, tagging through FPs is prolonged, yet when matched to QDs, genetic tags serve as the least cytotoxic fluorophores inside any cellular environment. The gene of interest is first cloned and then transformed into suitable cells. Once inside the cell, the bitterness due to FPs may be amplified, since they have their own unique proteinous feature which can lead to functional disorder of the attached protein of interest. In this direction, a striking effort [33] was made to genetically engineer one least cytotoxic red FP (FusionRed) for monitoring the protein dynamics in real time. In the chromophore surrounding region K69R and R203H mutations were transferred to mKate2.5, resulting in the dramatic pH stability of the protein. Earlier, mKate2.5 variant was yielded by inserting S132A, R164A, K182E and Y200N mutations along with the substituted C-terminus. In addition to pH stability, the final variant FusionRed exhibited photostability and maturation rate at par with other red-emitting fusions. Here we deliberate on the labeling schemes of immunofluorescent tags and genetic tags.
3.1. Labelling through immunofluorophores
There are two basic immunofluorescence techniques- direct and indirect. In former case, a single antibody conjugates directly with the fluorescent dye, whereas two different antibodies are exploited in latter- primary that recognizes target biomolecule and secondary that binds with fluorescent dye. However, for immunocytochemistry, direct labeling is carried out using both monoclonal and polyclonal antibodies. Background staining encountered with the use of secondary antibodies is also eliminated. In tissue sections, direct immunofluorescent technique (DIF) is principally applied in the identification of antibodies and other inflammatory proteins, to diagnose disease groups like pemphigus, lupus erythematosus, etc. that are histologically similar to separate under clinical investigations [34].
Additionally, labeled streptavidin binding (LSAB) and avidin-biotin complex (ABC) are the two common immuno-histochemical techniques that have been recently commercialized. In LSAB, primary antibodies are conjugated directly to fluorophores and detected by streptavidin [35] and in order to increase the spectral diversity for multiprotein analysis, antibodies are injected directly into live cells. Avidin-biotin labeling constitutes another receptor-ligand pair that works best in the secretory compartment. It is extremely useful for in vitro and histological investigations, however, for unknown reasons, this fluorophore toolbox is rarely explored in live cell imaging. pH measurements of various compartments in terms of different pKa values has been carried out through chicken avidin-biotin fluorescein conjugates, recombinantly expressed in different secretory compartments [[36], [37]]. This facilitates the study of mechanism of pH regulation in such secretory compartments. However, primarily the critical limitation includes the toxic nature of avidin-biotin fluorescein conjugate in cytosol and mitochondrial compartments or secondly avidin is saturated with biotin. G protein coupled receptors (GPCRs) regulate critical physiological functions through neurotransmitters, peptide hormones, etc. Therefore to visualize GPCRs in living or fixed cells, immunofluorescence is applied through two strategies. In one approach, through antibodies against the extracellular receptor regions, and in other case through an epitope tag [38]. Intracellularly sited receptors or receptor segments and epitope tags are recognized and visualized only after cell fixation and permeabilization (Fig. 3). The basic problem with immunolabelling is the larger size of immunofluorophores, interfering with multiprotein recognition. Furthermore, immunofluorescent techniques are limited to permeabilized cells, extracellular proteins, and intracellular proteins. Whenever a problem arises due to availability of low-grade antibodies, an epitope tag is applied to the target protein to express it in a recombinant manner.
 Fig. 3. In order to visualize GPCRs, how in non-permeabilized cells: (A) antibodies are used against extracellular receptor regions or N-terminal epitope tags. (B) In contrast, it’s only after cell permeabilization, that the visualization of intracellularly located receptors/segments or C-terminal epitope tags takes place.
Fig. 3. In order to visualize GPCRs, how in non-permeabilized cells: (A) antibodies are used against extracellular receptor regions or N-terminal epitope tags. (B) In contrast, it’s only after cell permeabilization, that the visualization of intracellularly located receptors/segments or C-terminal epitope tags takes place.3.2. Labelling through genetic tags
The major tagging hindrances have been overcome through genetically encoded FPs, which ensure perfect covalent tagging of FPs with the protein of interest. Genetic engineering followed by transformation procedure ensures an easier and perfect delivery of exogenous DNA into cells as compared to chemical dyes. By successfully developing different fusion constructs, FPs describes temporal dynamics of different metabolic processes in real time. For example, substrates binding to fusion constructs containing GFP undergo ubiquitin-proteasome dependent proteolysis [39]. FRET disruption by proteases emerged as a classical biophysical parameter for investigators searching for novel drugs against major lethal diseases such as AIDS and cancer. FRET based biosensors possess a protease-sensitive linker fused between a blue fluorescent protein (BFP) and green fluorescent protein (GFP). The disruption of FRET by proteolysis separate the donor and acceptor fluorophores and individual domains of metallothionein(MT) were generated by proteolysis of CdMT with substilin [40]. In exact sense the cleavage occurred between two lysine residues- 30 and 31 in the hinge region, ultimately cleaving the polypeptide chain between two fluorophores and thus in comparison to the control sample, energy transfer of dual-labeled MT decreased more significantly.
Cellular metabolite quantification in live cells constitutes the most important application of fluorescent proteins. For example, by inserting the peptide linker CaM and M13 between CFP and YFP, cameleons- the genetically encoded Ca2+sensors were constructed [26]. By increasing the intracellular levels of Ca2+ the affinity of CaM for the adjacent M13 sequence gets switched on. This causes a change in distance between two FPs that ultimately results into a large FRET. By replacing glutamate with glutamine residues in the pockets of Ca2+ −binding sites, the effective affinity of Ca2+for cameleons is regulated. Recently, well optimized FRET based nanosensors have been developed for measuring Ras and Rap1 activity [41] and imaging glutamate levels in brain [42]. We have successfully constructed special FRET based nanosensors for quantification of leucine [43] and methionine [44], for in vivo monitoring of zinc concentrations [45], lysine flux [46] and glycine betaine levels [47] and vitamin B12 levels [48].
4. Protein expression and localization studies
Fluorescent flow cytometry facilitates the visualization of endogenous proteins in their active state, very important for single cell profiling. However, immunofluorescent tagging is most suitable for endogenous proteins. To visualize the expression of particular proteins in situ within different cell lineages or individual cells, mutant phenotypes can be analyzed by immunofluorescence staining [49]. Although due to ample brightness of QDs, the detection limit advances much more and hence multi-label separation is achieved by varying spectral features. Though at light microscopic (LM) and electron microscopic levels (EM), QDs serve as appropriate tools for detection of proteins, however, it is only with the aid of electron microscopy that protein localization is effectively accomplished in subcellular structures. QDs are promising in electron microscopic observations, due to their size and electron dense core, and additionally, visualization is enhanced by silver staining. Recently, QDs have been utilized directly for concurrent immunolabelling of several endogenous proteins to acquire a correlated LM-EM imagery of cells [50]. Before proceeding for EM examination studies, we can swiftly get a precise analysis in LM by utilizing diverse range of QD labels. The fluorescent QDs at EM level overcome the need of antibody labeling, for example streptavidin labeled QDs detect surface proteins by using enzyme biotin ligase [51]. Correlated microscopy using tetracysteine ensures the preservation of ultrastructure against the immunolabelled microscopic imagery that needs permeabilization and makes inflexible fixation. Both FPs and immunofluorescent tags are extensively applied for subcellular allotment of proteins and there is a high correlation between live-cell and fixed localization investigations. Researchers utilize finest validated antibodies and suitable fixation procedures, critical to obtain better epitope accessibility and an accurate protein distribution in vivo.In terms of cellular and subcellular localization however, FRET based nanosensors are comparatively more efficient than all other labeling methods. Compared to adding chemical dyes exogenously, such nanosensors can be expressed within stable cell lines and transgenic animals. The strategic approach is combination of FRET with metabolite recognition capability of a bacterial periplasmic binding protein (PBP) [52], followed by successful transfection of resultant nanosensor into any cell type. This expands the possibilities for high-throughput screening, rigorous cell, developmental and physiological studies. In case of plants, most cells contain a large vacuole in their cytosol making it difficult to quantify plant metabolite concentrations in a given compartment in the little available cytoplasmic space. The FRET based nanosensors are comfortably targeted to sub-cellular locations, facilitating the high-resolution mapping of signals within plant cells. Since there is an enormous range of metabolites in plants with highly complicated transport pathways that are quite contrasting against the animal systems.
5. Visualization of protein activity in live cells
5.1. Monitoring protein conformational changes and protein–protein interactions
A healthier spatiotemporal resolution of protein conformational changes is usually achieved by fixing a protein domain in between two FPs- Cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) respectively. This is followed by FRET between two FPs, as the ligand binds with ligand sensing domain. Protein that connects the two FPs is engineered to undergo conformational changes in reaction to very important signals. With the aid of an appropriate localization signal, the resultant biosensors are targeted to precise subcellular compartments. A wide range of FRET based nanosensors have been constructed for measuring various metabolites, to monitor the activity of many proteases, check the balance between kinases and phosphatases, for sensing neurotransmitters and other metabolites [53]. FRET efficiency varies with distance and orientation between donor and acceptor FPs. However, circular permutation of any of the two FPs [54] or slight adjustments in the length of linker region greatly alters the FRET, as a result the frequent crop up of FRET responses are more due to reorientation and less by means of change in their inter fluorophore distance.
In living cells, FRET visualizes the dynamic protein–protein interaction, by means of two FP tagged proteins, whence a ligand binds with the ligand binding domain (Fig. 4). FRET processing occurs as the two FPs are within the intermolecular distance of 6–8 nm. However, recent approaches extended to the three FP tagged FRET based biosensors. It was carried out by valuable addition of monomeric red fluorescent protein (mRFP) to the pair of CFP/YFP. Such trimeric FRET based nanosensors again endow with deeper resolutions. In such FRET experiments, CFP acts as a donor for YFP and in turn, YFP as the next donor for mRFP (Fig. 5). On the basis of same principle, bimolecular fluorescence complementation (BiFC) is low pace chromophoric interaction between two split fragments of a FP (Fig. 6) fused with two interacting proteins. Fluorescence correlation spectroscopy works in a novel way- using the two different tags aided with two different colors that interact and diffuse as a pair [55]. There are simple colocalization events that indicate substantial protein–protein interactions. As seen in case of protein kinase A (Fig. 7), its regulatory subunit and catalytic subunit are co-expressed. The latter is fluorescently tagged and destined to plasma membrane, and whence the adenosine 3′, 5′ −monophosphate (cAMP) levels are increased, the two partners get dissociated, and therefore FRET visualization takes place.
 Fig. 4. FRET phenomenon: FRET takes place when ligand binds with the ligand sensing domain, through which CFP (donor) and YFP (acceptor) are in proximity.
Fig. 4. FRET phenomenon: FRET takes place when ligand binds with the ligand sensing domain, through which CFP (donor) and YFP (acceptor) are in proximity. Fig. 5. Three fluorophore based FRET process: by the attachment of gene 1 with gene 2, intermolecular FRET occurs first between CFP and YFP. Afterwards, gene 3 encoding RFP also gets attached, which acts as new acceptor fluorophore for another FRET.
Fig. 5. Three fluorophore based FRET process: by the attachment of gene 1 with gene 2, intermolecular FRET occurs first between CFP and YFP. Afterwards, gene 3 encoding RFP also gets attached, which acts as new acceptor fluorophore for another FRET. Fig. 6. BiFC leading to the formation of a functional chromophore. Before this process, the two GFP halves are separate and BiFC leading to their proper folding resulting into the formation of a functional chromophore.
Fig. 6. BiFC leading to the formation of a functional chromophore. Before this process, the two GFP halves are separate and BiFC leading to their proper folding resulting into the formation of a functional chromophore. Fig. 7. Indirect FRET: Addition of a phosphate group to the substrate by Kinase enzyme leads to a conformational change that brings CFP and YFP close together, which ultimately results into FRET.
Fig. 7. Indirect FRET: Addition of a phosphate group to the substrate by Kinase enzyme leads to a conformational change that brings CFP and YFP close together, which ultimately results into FRET.Halotags have been used for FRET technique and it has been validated that compared to other bio-conjugated dyes, utilizing the Halotag protein linked with enhanced green fluorescent protein (GFP) displayed superior fluorescence stability [56]. Halotags have been applied to monitor protein–protein and protein-DNA interactions. They have been used to monitor the interaction of bromodomain protein (BRD4) and histone deacetylase (HDAC1) along with other additional proteins [57]. Halotag has been adapted for the investigation of epidermal growth factor receptor Ras-extracellular signal regulated kinase (ERK) mitogen activated protein (MAP) kinase pathway in living cells. FRET has been further adapted for Halotag mediated conjugation and site specific decoration of molecular beacons, utilizing two different FP fusions, thus enabling easy detection of target nucleotide sequences [58]. In live cells, the protein–protein interactions and alternative protein conformations have been detected through bipartite tetracysteine display. Such profluorescent biarsenical reagents- FlAsH-EDT2 and ReAsH-EDT2 have led to detection of early protein misfolding events associated with Alzheimer’s and Parkinson’s diseases and for high-throughput screening of compounds that stabilize discrete protein folds[59].
5.2. Visualizing protein dynamics
Protein labeling enables us to track its redistribution within a cell by using various imaging techniques which facilitate visuals of protein dynamics. Imaging readouts are therefore sought for such translocation processes that includes- monitoring buildup of polyphosphoinositides through FP-tagging of pleckistrin homology domains, proteins diffusing at steady state rates or involved in inter-compartmental exchange. Intensity fluctuations are statistically interpreted in fluorescence correlation spectroscopy. These fluctuations are because of fluorescent object mobilities, focused in front of a laser. Correlation of such images gained through spectroscopy efficiently measures these fluorescent variations. This facilitates the easy way illustration of mobility and interfacings of labeled molecules in real time. The critical factor for photomarking method is photochemical sensitivity. It involves dequenching of fluorophores, either through photoactivation (Fig. 8) [60] or FP destruction. These two photo-marking modifications in fluorophores lead to the imaging of participant proteins. However, it needs to be emphasized, that the role of large fluorescent tag sizes is critical for passive mode of translocation.
 Fig. 8. Showing the process of photoactivation, in which intense illumination causes the quenching of a fluorescent protein, changing its spectrum. This is followed by movement of fluorophores towards each other.
Fig. 8. Showing the process of photoactivation, in which intense illumination causes the quenching of a fluorescent protein, changing its spectrum. This is followed by movement of fluorophores towards each other.This technique sees the tracking down of unimolecular protein translocations. It harvests the dual only fluorescent behavior (both poor and bright fluorescence in molecules or organelles) and uses automated software to track visualizations in the form of single videos. In the macromolecular structures of filamentous protein aggregates like actins and microtubules, upon fluorescent tagging the whole protein turnover processing is echoed by dynamic translocation of fluorescent patches. QDs were conjugated with EGF that resulted in specific co-localization with ErbB1-GFP chimeric receptor [61]. It was revealed that against activated ErbB1, Cy5-conjugated antibodies determined the activation of the receptor. Colocalization of transferrin labeled with Alexa fluor revealed that the complex internalization was a clathrin dependent phenomenon. By utilizing, distinctly synthesized EGF-QD, it was found that receptor oligomerization lead to retrograde transportation of receptor bounded single EGF-QD. It was furthermore revealed by the novel phenomenon of fluorescent recovery after photo bleaching (FRAP) that both actin flow and retrograde receptor transport occurred simultaneously (Fig. 9). With the use of biotin Alexa, the specific quenching of EGF-QDs revealed that receptor internalization occurred at cell body and not on filopodia. Henceforth, the involvement of filopodia to transport the activated receptors to cell body is brightly persuasive.