1. Introduction
The genetic transformation in plants has revolutionized agriculture by facilitating the introduction of foreign genes into the agronomically and horticulturally important specie. This technology leads to the expression of novel traits such as pest resistance, disease resistance, and quality improvement. The transgenic plants are generated based on the genetic transformation techniques mediated by Agrobacterium tumefaciens, particle bombardment, and DNA uptake into protoplast. The transgene integration, mediated by these techniques takes place at random sites in the plant genome. The position of genomic integration and the complexity of the integrated DNA influence the level of transgene expression [58], [39], [21]. Also, the transgenes inserted at random positions may lead to redundant mutations because of its insertion in the active plant genes [37]. The development of techniques that mediate transfer and integration of the foreign genes at specific pre-determined locations obviates many complications associated with the existing gene transfer methods. The introduction of foreign genes via Gene Targeting (GT), which is based on the Homologous Recombination (HR), offers many advantages such as precision gene integration, single copy transgene insertion, and high expression of the transgenes. It allows the construction of ‘safer’ transgenic crops, with no unknown ‘position’ effects due to random integration.
GT is a genetic technique that uses HR to alter a specific DNA sequence in an endogenous gene at its original locus in the genome. Paszkowski et al. [56]integrated antibiotic-tolerant gene into the tobacco genome using GT originally. Various HR-dependent approaches have also successfully targeted genes in plants [33], [75], [80].
In this review, we systematically reviewed the performance of various methods and approaches about the introduction of plant gene targeting. The HR, site-specific recombination, Zinc Finger Nucleases (ZFNs), Transcription Activator-Like Effector Nucleases (TALENs), and clustered regularly interspaced short palindrome repeats (CRISPR) has taken into consideration.
2. Genome editing tools
2.1. Homologous recombination
HR is a DNA maintenance pathway that protects the chromosomes against damages involves both the DNA strands, such as Double Strand Breaks (DSBs) and interstrand crosslinks. HR, illegitimate recombination or Non-Homologous End Joining (NHEJ), and Single-Strand Annealing (SSA) are the three different ways of foreign DNA integration into the native genome. SSA, the third path of repair, requires the presence of repeated sequences on both sides of a break. After the exonuclease degradation of the 5′ ends, repair occurs by annealing of the two complementary sequences, which leads to the loss of the geneticinformation contained between these repeats.
Puchta [63] reported that one of the most efficient and effective means of improving the frequency of HR frequency to develop a break in the chromosome at the target site. The repair mechanism originates at the break by simulating the cell’s DNA system, and the homologous template proceeds through the HR during the repair process. GT has been widely used in mice and yeast, and its efficiency in plants was not sufficient for the routine applications [60], [24]. Different methods were tested for increasing the GT efficiency in plants. Moreover, DSBs are created by a rare enzyme, I-SeeI, which could improve the homologous integration frequency at the target site [61]. Such strategy entails transgenic target sites that are randomly inserted into the genome, and thus, it would not be likely to target endogenous genes [62], [25], [59] referred that rad9 and rad17 mutants lead to the induction of high HR frequency.
Several attempts have been made to enhance the competitiveness of the HR machinery by expression of the heterogeneous factors in other organisms. RecADNA recombination protein in Escherichia coli has improved the frequency of intrachromosomal recombination [64]. Further, Reiss et al. [64] stated that the line of RecA- overproduction strategy did not modify the GT frequency. Provided the evolutionary conservation of mechanisms in recombination, the presence of complete sequences of Rice and Arabidopsis characterized endogenous regulatory components of HR/NHEJ [50] have applied forward genetic screens for the genotoxic treatments of the affected mutants, indicating the original components that influenced HR levels. The continuation of such efforts would assist in the understanding of the circuits of plant-particular regulation that are included in the repairing of DSB [26].
Previously, the frequency of the HR-dependent GT was identified in the order of 10−3 and 10−6 with regard to the random combination of GT vector [28], [40], [79], [7], [66], [23], [24], [65], [32]. Subsequently, it was found that the induction of DSBs maximized the HR frequency by certain magnitudinal orders. Therefore, engineered nucleases were developed as an appropriate method for enhancing the efficiency and successful deployment of GT in mammals that maximized gradually as illustrated by [36]. Voytas [80] stated that DSBs induction at specific genome locations was effective in enhancing the efficiency of GT in plants.
Genome editing, or genome editing with engineered nucleases (GEEN) is a type of genetic engineering to insert, delete or replace DNA in the genome of an organism using engineered nucleases, or “molecular scissors.” These nucleases create site-specific double-strand breaks (DSBs) at desired locations in the genome. The induced DSBs are repaired by non homologous end-joining (NHEJ) or homologous recombination (HR), resulting in targeted mutations.
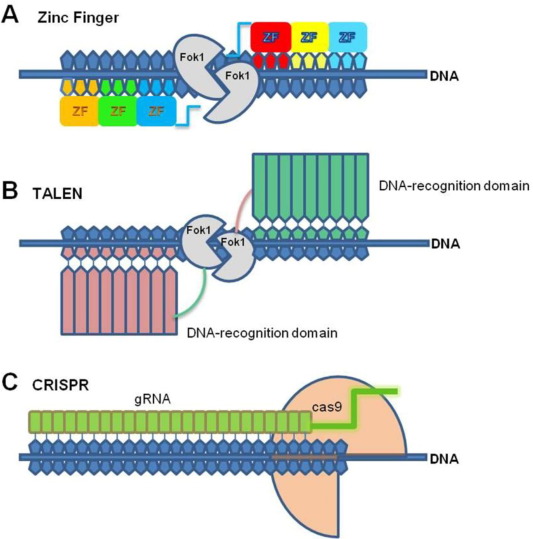
There are currently 3 types of engineered nucleases being used: zinc finger nucleases (ZFNs), transcription activator-like effector-based nucleases (TALEN), and the CRISPR-Cas system. Fig. 1 illustrates the structure and mode of action of these nucleases and Table 1 shows the comparison between these technologies.
 Fig. 1. Schematic representation of Genome Editing Tools.
Fig. 1. Schematic representation of Genome Editing Tools.Table 1. Comparison of Genome Editing Technologies.
| Activity | ZFN | TALEN | CRISPR |
|---|---|---|---|
| Recognition | Protein-DNA | Protein-DNA | RNA-DNA |
| DNA targeting specificity determinant | Zinc-finger proteins | Transcription activator-like effectors | RNA |
| Nuclease | Fok1 | Fok1 | Cas9 |
| Target sequences | 2 × 12 nucleotide and up | 2 × 16 nucleotide and up | Nearly 20 nucleotide |
| Construct size | (1 kb) 2 | (3 kb) 2 | 4.2 kb (Cas9) + 0.1 kb (RNA) |
| Construct | Zinc finger sequence specifically recognizing 3 bp sequence linked to Fok1 | Protein sequence specific to binding a nucleotide sequence linked to Fok1 | A 20nt crRNA fused to tracrRNA and Cas9 endonuclease |
| Construct | Zinc finger sequence specifically recognizing 3 bp sequence linked to Fok1 | Protein sequence specific to binding a nucleotide sequence linked to Fok1 | A 20nt crRNA fused to tracrRNA and Cas9 endonuclease |
| Cost and time involved in assembly | Very expensive and time consuming | Relative expensive and time consuming | Low cost and minimum time |
| Multiplexing | No | No | Capable |
| Success Rate | Low | High | High |
2.2. Zinc finger nucleases
ZFNs are restriction enzymes with Zinc Finger (ZF) domains that recognize a particular sequence of DNA, fused to the nuclease domain of restriction enzyme Fokl [38]. Since the domain of ZF could be engineered to focus on novel sequences of DNA, ZFNs were exploited to engineer the endogeneous genome loci, particularly in the eukaryotic systems, [10]. According to [45], in the case of ZFNs, one module of DNA-binding involves nearly 30 amino acids and identifies 3 nucleotides integrating module of DNA-binding. It allows the recognition of 9–18 bps of DNA sequences. ZFNs were used for creating breaks in the site-specific chromosome, particularly in the absence of pre-engineered sites for the target [3], [76].
The development of ZFN-mediated GT provides Molecular Biologists with the ability to modify the plant genomes site-specific and permanent via homology-directed repair of a targeted genomic DSB. ZFNs can be used to induce DSBs in specific DNA sequences and thereby promote site-specific HR and targeted genomic manipulation. ZFNs have a DNA recognition domain that involves an array of Cys2-His2 ZF. ZFs recognize and bind to particular nucleotide triplets. Various ZFs can be combined together for generating DNA-binding arrays that would identify the expanded sequence patterns with high affinity and greater specificity [15], [55], and [68]. The gene constructs were made from the custom-designed ZFNs which were designed to cut at specific DNA sequences at a preselected locus in the plant genome. This was due to the efficiency and directiveness of the ZFs for a broad range of DNA sequences. A site-specific ZF endonuclease has been successfully employed to induce site-specific mutations by Non-Homologous End Joining in Arabidopsis [46].
By convention, the targeted genome modification (TGM) was frequently performed using the synthetic domain of ZFNs that is fused to a cleavage domain of Fok1 [77], [11]. ZFNs were used to modify endogenous genes in various organisms, cell types, and plant species including Arabidopsis [54], [82], soybean [18], maize [70] and tobacco [74]. Most of the constraints on the application of ZFN encompass the limited quantity of existing target sites, more effects on context dependence between the low targeting specificity and efficiency, repeat units and the effects of frequent off-target caused partially by the non-specific binding of DNA [20].
In Drosophila, the engineered ZFNs have identified the yellow gene that was observed in the larvae in the presence of the donor DNA. It was either joined elsewhere in Drosophila genome or seen as free-floating molecules released from the chromosomes of Drosophila by FLP recombinase [3].
Bozas et al. [6] studied the genetic analysis of ZFNs-induced GT in Drosophila. Using ZFNs for cleaving the target in the chromosome, high frequencies of GT in the germ line of Drosophila was targeted. Both local mutagenesis through NHEJ and replacement of the gene through HR are simulated by targeting the cleavage. In this review, we focused on the mechanism related to the processes of applying materials for ry or rosy locus. HR-dropped frequency was significant in homozygous flies particularly for mutations in okr (Rad54) or spnA (Rad51) genes, and two components of invasion-mediated Synthesis-Dependent Strand Annealing (SDSA) pathway. When SSA was blocked by using circular donor DNA, HR was fully removed. This further show that the majority of the NHEG products, were produced in a lig4- dependent process. When both lig-4 and spnA were mutated and provided with a circular donor, the frequency of rymutations was still high, and no products of HR could be observed. Further, it was stated that the local mutations given in such circumstances required an alternative, like lig-4 mechanism, for independent end-joining. These outcomes indicate the types of repair operating pathways for DSB in the mentioned GT systems. It was also found that the results could be biased toward gene replacement by disabling the main pathway of NHEJ and moving toward simple mutagenesis by interfering with the HR process.
2.3. Transcription activator-like effector nucleases
TALENs have been developed as an alternative to ZFNs, particularly for targeted genome modification, and have indicated high capability for precise manipulation of the genome [16] Similar to ZFNs, TALENs comprise an engineered oriented domain of TALE (Transcription Activator-Like Effectors) DNA binding and cleavage domain of Fok1. The customizable binding domain of TALE DNA consisted of certain approximately identical repeat arrays in tandem that could target any provided sequence based on simple RVD (repeat variable di-residue) code for nucleotide recognition [4], [5]. In the recent past, the modification of TALEN-mediated genome was widely accepted in yeast [43], rat [73], [72], fruit fly [4], human pluripotent and somatic cells [51], [27], nematode [81], livestock [9], silkworm [47], plants [69], [48], [83], zebrafish [67], [30], [2], [8], [19], [52], Xenopus embryo [41], and many other organisms.
TALENs are the integration of the cleavage domain of Fokl and binding domains of DNA derived from the TALE proteins. TALEs are the naturally occurring proteins from the plant genus of pathogenic bacteria namely Xanthomonas. [83] pointed out the methods of plant genome targeted modification using TALENs. Further, it was stated that the methods were optimized using protoplasts of Nicotiana tabacum targeting the acetolactate synthase gene. TALENs comprise DNA-binding domains involving in a series of 33–35 amino acid repeats, with each domain recognizing a single base pair. Therefore, a minimum of only 4 types of the module in DNA-binding are required for recognizing C, T, A, and G. Also, it was noted that the single-base identification of TALE-DNA binding domain repeats provided greater flexibility than design when compared to the triplet-restricted ZFNs as mentioned by [44]. Thus, it could be stated that TALENs are physically larger when compared to ZFNs by identifying the similar number of nucleotides.
TALENs have evolved as the reagent of choice for effectively changing the eukaryotic genomes in a targeted fashion [44], [73], [1]. Although TALENs have been shown to perform at high efficiency in many human cell lines and animal species, but its use in plant genome modification was shown only in 3 species (rice, tobacco and Arabidopsis). Moreover, various studies have used TALENs for creating mutations, especially for NHEJ [13], [4], [8], [42].
It has been shown that the transient assay in the protoplast is an accurate, rapid, and reliable method for assessing the nuclease activity in tobacco and Arabidopsis [82], [83]. The repeat arrays in TALEN were 1st cloned into the expression vector pZHY051, for assessing the nuclease activity in the protoplasts. The transient assay in the protoplast was developed in both Brachypodium and rice. Encoding constructs for TALEN were introduced into the protoplasts using Polyethylene glycol (PEG). After two days of incubation, the genomic DNA was developed for each sample, and DNA fragments entailed each target site that was amplified by the PCR. The PCR products were digested with restriction enzymes and separated by agarose gel electrophoresis. The PCR amplicons were cloned into the T-A cloning vector, and nearly 30–50 individual clones were examined for mutations by DNA sequencing. TALEN-induced mutations frequencies at each target site in endogenous protoplasts were analyzed. The positive correlations of the nuclease activities in the protoplasts and changed calli in tobacco have been also discussed as illustrated by [83].
2.4. CRISPR/Cas
Targeted Genome Engineering (TGE) was developed as an alternative to traditional transgenic and plant breeding methods for enhancing productivity and ensuring sustainable production. TALENs and ZFNs were used, particularly in genome mutation at specific loci, but such systems need 2 novel binding proteins for DNA flanking a sequence of choice with a C-terminal module for FokI nuclease. Thus, such methods have been mostly accepted by the plant research community. Further, a new method was developed to enhance the efficiency of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated (Cas) type II prokaryotic system as an alternative for genome engineering [71]. Various studies mentioned that the ability for reprogramming specificity of CRISPR/Cas endonuclease using customizable small non-coding RNAs had formed the stage for editing applications of a novel genome [49], [12], [31], [14], [35], [34]. The system is based on Cas9 nuclease and an engineered single guide RNA (sgRNA) that gives a targeted sequence for nucleic acid [12], [34], [14]. According to [83], similar to TALENs and ZFNs, CRISPR technology has become one of the new plant breeding techniques. Such techniques make it feasible to introduce modifications in the plant genome that are non-distinguishable from those introduced by the traditional breeding and physical or chemical mutagenesis [14].
In plants, CRISPR/Cas9 system was deployed using the transient expressionsystems that enabled rapid optimization and execution of the method. The applied transient assays in plant research are protoplast and leaf tissue transformation using the method of agro-infiltration. Both methods were used for sgRNA and Cas9. The benefit of employing the protoplast strategy is the probability of achieving high gene co-expression gene even from the isolate plasmids. The separation of protoplasts from the plant tissue needs enzymatic digestion and cell wall elimination. The procedure could be time-consuming as the protoplast cultures are prone and fragile to contamination. An alternative procedure is the agro-infiltration assay that functions on whole plants as well as it is time saving. Such system works by A. tumefaciens strains infiltration carrying a binary plasmid that is comprised of candidate genes for expressing. Gene co-expression efficiency by agro-infiltration assay was lower when compared with the protoplasts, and integrating various genes of choice in one vector is possible.
Target specificity is a significant problem for all technologies for genome editing encompassing CRISPR/Cas. Numerous studies have analyzed the specificity of CRISPR/Cas system in human cells and in vitro [10], [22], [29], [49], [57]. The major outcome is that 3′ end of the guide sequence inside the sgRNA particularly indicates the target specificity of CRISPR/Cas system. This result is consistent with the previous studies by other researchers [35], [17], [14]. The mismatches between the guide sequence and DNA target of the sgRNA situated inside the final 8–10 bp of 20 bp sequence in target mostly remove the recognition of the target by Cas9 whereas mismatches towards the 5′ target end are better tolerated [17], [53].
3. Conclusion
In this study, we discussed various approaches and methods regarding GT. The extensive growth and development in the plant GT have been summarized. This review indicates the significance of this research in developing the plant GT methods.
The reviewed findings indicate that one of the most efficient means for improving the HR frequency is to develop a break in the chromosome at the target site. The use of SSR gives the most straightforward method for chromosome excision. The removal of the marker genes from genetically modified and commercial plants is of specific interest because it would deliver a new generation of products for transgenic plants. It was noticed that ZFNs were exploited particularly in the eukaryotic systems to engineer endogenous genome loci. The TALENs have been developed as the reagent of choice for effectively changing eukaryotes genomes. They have a high efficiency in many human cell lines and animal species, whereas only 3 plant species showed modification due to TALENs. The CRISPR technology is also one of new plant breeding techniques.
The difficulty in employing the transformation techniques in the case of higher plants at a higher frequency in order to get GT events has not been resolved yet. Arabidopsis plants are readily transformed by dipping plants with Agrobacterium strains carrying the transgene. This simple procedure has not been applied successfully in major crop species yet. The efforts required for transforming and screening higher plants for GT events are tedious. The development of the GT technology represents a crucial step in improving our understanding of single gene functions in its genome background by gene knocking. Moreover, it has the potential to increase the public acceptance of the plant gene modification by molecular techniques.