Recurrent chromosomal deletions are frequently associated with human diseases. Hematologic malignancies, in particular, often harbor focal deletions, most commonly in the hemizygous state. These may drive disease by providing one of two hits toward complete gene inactivation or through haplo-insufficiency of one or more genes. Deletion of the long arm of chromosome 7q (del7q) is a common cytogenetic abnormality in patients with hematologic malignancies, particularly myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). Because of its frequency and strong association with adverse prognosis, several efforts have been made over the years to map the critical chr7q region with physical mapping approaches aimed at pinpointing minimal regions of overlap from patients with myeloid malignancies harboring different chr7q deletions 1, 2, 3. Although methods of detection of increasingly higher resolution have become available over the years (from G-banding to next-generation sequencing), physical mapping is limited by the number of informative cases. The total number of existing patients with del7q-MDS is limited, and the majority of them harbor very large deletions encompassing the whole chromosome, or the entire long arm, or regions several megabases in length that contain hundreds of genes 1, 2, 3. Furthermore, in the context of complex karyotype MDS and AML, del7q is hard to evaluate, and so are cases of loss of heterozygosity that may be copy number neutral.
We previously provided proof of principle of a functional mapping approach that allowed us to pinpoint a critical chr7q region using human induced pluripotent stem cells (iPSCs) with chr7q deletions derived through reprogramming of MDS patient cells and through genetic engineering, using a chromosome elimination cassette consisting of two loxP sites in inverse orientation to one another inserted into chr7q [4]. However this approach was severely limited, as it relied on the generation of random deletions that were selected with the herpes simplex virus thymidine kinase (HSV-tk) gene/ganciclovir system, via a HSV-tk transgene inserted in a near-telomeric region of chr7q in a prior genetic engineering step [4]. The limitations of this approach thus were, first, the lack of control over the exact length and borders of each deletion and, second, the rarity of informative clones with unique deletions.
We thus set to develop strategies that enable the generation of targeted deletions and afford control over the chromosomal boundaries. Deletions of megabase scale up to 15 Mb have previously only been generated in cancer cell lines by using various programmable nuclease technologies (zinc finger nucleases, TALENs, or CRISPR) 5, 6, 7. A 30-Mb deletion has also been generated through CRISPR in a near-haploid human cell line (HAP1) [8]. Induction of double-strand breaks (DSBs) by programmable nucleases has also been exploited to generate large genomic rearrangements, such as duplications and inversions in human cancer cell lines [9,10], as well as to eliminate whole chromosomes in human cell lines or mouse ESCs (Y chromosome or chr21 in trisomic cells) [11,12]. The largest engineered deletion reported in normal human cells so far is a 725-kb deletion engineered in Duchenne muscular dystrophy (DMD) iPSCs for disease correction [13].
Here, we describe the successful generation of targeted megabase-scale deletions in normal human iPSCs. We report that single or double Cas9-mediated DNA DSBs can enable the isolation of clonal human iPSC lines with targeted terminal and interstitial deletions as large as 22 Mb. This is the first demonstration that targeted deletions at the megabase range can be engineered in normal diploid human cells.
Methods
Human iPSC culture
Human iPSCs were cultured on mouse embryonic fibroblasts (MEFs) or under feeder-free conditions as previously described [4].
AAV-mediated gene targeting
The 5′ and 3′ homology arms of the acylglycerol kinase–adeno-associated viral (AGK–AAV) vector consisted of chromosome 7 nucleotides 141,554,358–141,555,466 and 141,555,467–141,556,540 (hg38 human genome assembly), respectively. The entire sequence was amplified from isogenic human genomic DNA in two independent polymerase chain reactions (PCRs), cloned, and sequenced, and one allele was chosen for subsequent amplification and cloning. AAV vector production was performed as described [4]. Two hundred thousand iPSCs were transduced under feeder-free conditions, replated on neomycin-resistant MEFs, and selected with 0.5 μg/mL G418 for 5–7 days. Eighteen single-cell clones were screened by PCR with the primers 5′-F (CAGCTACTTGGGAGGCTGAG), 5′-R (ATACTTTCTCGGCAGGAGCA), 3′-F (TCGCCTTCTTGACGAGTTCT), and 3′-R (AAATGCCAGGGAGACATGAG).
Engineering chromosomal deletions with Cre-loxP recombination
Transduction with a Cre recombinase-expressing integrase-deficient lentiviral vector was done as previously described [4]. Ganciclovir selection was performed at a concentration of 200 μmol/L for 10–15 days. Twenty ganciclovir-resistant clones were screened by quantitative PCR, probing different regions along the length of chromosome 7 (Supplementary Table E1, online only, available at www.exphem.com). PCR of the junction with primers 7q34 F (TAGGAAGGGAGCTTCCAGGT) and 7q36.3 R (CTCCAAGGACAAACACGTTG) and Sanger sequencing were used to confirm the predicted recombined sequence.
CRISPR/Cas9-mediated deletions
To generate the C(35-qter)13, C(q35-36.3)8, and C(q33-36.3)5 clones, three gRNAs were designed targeting the 7q35, 7q36.3, and 7q33 genomic regions, respectively. Specifically, gRNAs 7q35 (GCAGAGCAGTGACGGTAAGCGGG), 7q36.3 (GCCGGACGCCGTTCTCCGCGAGG), and 7q33 (GTCTAACCTCATTGGACCGGAGG) were amplified in a two-step overlapping PCR downstream of the U6 promoter sequence and cloned in the gRNA/Cas9 lentiviral plasmid, co-expressing a human codon-optimized Cas9 with a nuclear localization signal (from George Church, Addgene Plasmid No. 41815), linked to mCitrine by a P2A peptide, driven by the CMV promoter, and gRNA driven by the U6 promoter [14]. The vector was packaged as described [4].
The N-2.12-D line, harboring a HSV-tk transgene for negative selection inserted in chr7q36.3 [4] was dissociated with accutase. Two hundred fifty thousand cells were plated on Matrigel. The next day, the cells were transduced with two gRNA/Cas9 lentiviral vectors (targeting 7q35 and 7q36.3 or 7q33 and 7q36.3) at high multiplicity of infection (MOI). Two days later, the cells were dissociated into single cells with accutase and replated on MEFs. A portion of cells were used for flow cytometry to evaluate transduction efficiency by mCitrine expression. Ganciclovir selection was performed at a concentration of 200 μmol/L for 10–15 days. All ganciclovir-resistant colonies (17 and 15, respectively) were picked in separate wells of a six-well plate, allowed to grow for approximately 3–6 days, and screened by qPCR, probing different regions along the length of chromosome 7 (Supplementary Table E1). One clone from each condition was selected and expanded for further analysis. The junction sequences in clones C(35–36)8 and C(33–36)5 were PCR-amplified with primers junc-7q35-36.3-F (TAAATGCCTTGGCATGAACA), junc-7q35-36.3-R (CGCTGAAAGCTACGAAAAGG), junc-7q33-36.3-F (GGGAGCAACTGGATGTGTTT), and junc-7q33-36.3-R GAAGGCACGAGAAGGTGAAG and sequenced.
Array comparative genomic hybridization
Array comparative genomic hybridization (aCGH) was performed using Agilent's GenetiSure Cancer Research array CGH + SNP (2 × 400k) according to the manufacturer's instructions (Agilent Technologies, Santa Clara, CA). Test and reference DNA were digested with AluI and RsaI (Agilent) and labeled with Cy5-dUTP and Cy3-dUTP (Agilent), respectively. Human female reference DNA provided in Agilent's SureTag Complete DNA labeling kit was used as a sex-matched control. The microarray was hybridized in a SureHyb oven (Agilent) at 65°C for 48 hours. Posthybridization washes included Agilent Oligo CGH Wash Buffer 1, Agilent Oligo CGH Wash Buffer 2, and acetronitrile. The arrays were scanned using Agilent's G2600D scanner, and image analysis was performed using the default settings of the Feature Extraction software Version 11.5.1.1. Agilent Cytogenomic Version 4.0.3 software was used to visualize, detect, and analyze genomic gains and losses using the ADM-2 algorithm.
Hematopoietic differentiation
For hematopoietic differentiation, spin embryoid bodies (EBs) were prepared and cultured in APEL medium, as described [15]. Briefly, cells were dissociated into single cells with accutase and plated at 3,000 cells per well in round-bottom low-attachment 96-well plates in APEL medium containing 30 ng/mL bone morphogenetic protein 4 (BMP4) and 10 nmol/L Y-27632. The plates were centrifuged at 800 rpm for 5 min to induce EB aggregation. After 24 hours, the medium was replaced by APEL medium containing 30 ng/mL BMP4 and 50 ng/mL FGF2. After 2 days, the cytokine cocktail was changed to 20 ng/mL vascular endothelial growth factor (VEGF), 10 ng/mL FGF2, 100 ng/mL stem cell factor (SCF), 20 ng/mL Flt3 ligand (Flt3L), 20 ng/mL thrombopoietin (TPO), and 40 ng/mL IL-3. At day 8, EBs were collected and resuspended in Stem Pro34 SFM medium with 1% nonessential amino acids (NEAAs), 1 mmol/L L-glutamine, and 0.1 mmol/L 2-mercaptoethanol (2ME), supplemented with 100 ng/mL SCF, 20 ng/mL Flt3L, 20 ng/mL TPO, and 40 ng/mL IL-3. The medium was thereafter replaced every 2 days. At the end of the EB differentiation culture (day 14), the cells were collected and dissociated with accutase into single cells for flow cytometry and colony assays.
Flow cytometry
The antibodies CD34-PE (clone 563, BD Pharmingen) and CD45-APC (clone HI30, BD PharMingen) were used. Cell viability was assessed with DAPI (Life Technologies). Cells were then assayed on a BD Fortessa, and data were analyzed with FlowJo software (Tree Star).
Clonogenic assays
For methylcellulose assays, the cells were resuspended in StemPro-34 SFM medium at a concentration of 3 × 104/mL. Five hundred microliters of cell suspension was mixed with 2.5 mL MethoCult GF+ (H4435, Stem Cell Technologies), and 1 mL was plated in duplicate 35-mm dishes. Colonies were scored after 14 days and averaged between the duplicate dishes.
Gene expression analysis by qRT-PCR
RNA was isolated from N-2.12-D, C(35-qter), C(35–36)8, and C(33–36)5 with Trizol (Life Technologies). Reverse transcription (RT) was performed with Superscript III (Life Technologies), and qPCR was performed with the SsoFast EvaGreen Supermix (Bio-Rad) using primers for Cas9 with sequences F (CATATCGTGCCCCAGTCTTT) and R (CCGTTGTGTGATCAGTTTGG). Reactions were performed in triplicate in a 7500 Fast Real-Time PCR System (Applied Biosystems).
qPCR for chromosome 7 dosage
TaqMan qPCR was performed as previously described [16] with the primers and probes listed in Supplementary Table E1.
Results
Engineering a hemizygous 16-Mb chr7q deletion in iPSCs using Cre-loxP technology
We previously defined a 20-Mb region in chr7q that, when deleted, reproduced the phenotype of iPSCs derived from patients with del7q-MDS [4]. To engineer partial deletions within this previously defined 7q32.3–7q36.1 region, we first used a classic Cre-loxP approach. We used a previously generated normal iPSC line, in which a loxP site together with an HSV-tk transgene for negative selection was targeted into a near-telomeric region of chr7q, 7q36.3 (N-2.12-D) [4] (Figure 1A). Using AAV-mediated targeting, specifically ATG-trap of the AGK gene (selected on the basis of high expression in human iPSCs), we inserted a second loxP site together with a neomycin resistance (Neo) cassette into chr7q34. In this configuration, the two loxP sites are in the same orientation and at a distance of 16 Mb from each other (Figure 1A; Supplementary Figure E1 A,B, online only, available at www.exphem.org). Eight of 18 neomycin-resistant clones were found to be correctly targeted by PCR (Figure 1B). Clone 1 was used further and transduced with an integrase-deficient lentiviral vector to transiently express Cre recombinase (Supplementary Figure E1 B). After AAV transduction, 20 ganciclovir-resistant clones were screened by qPCR with multiple probes along the length of chromosome 7. One clone (AGK1Cre3) was found to harbor the predicted Cre recombinase-mediated deletion, 7q34–7q36.3, which was confirmed by aCGH (Figure 1C). The expected recombination event between the two loxP sites was confirmed by sequencing of the junction (Figure 1D). However, karyotypic analysis of clone AGK1Cre3 revealed clonal abnormalities (duplication of chromosome 5 and trisomy 12) (Supplementary Figure E1 C). These were likely random chromosomal abnormalities and not mediated by Cre recombinase. However, given their clonal nature, clone AGK1Cre3 was not used for any further experiments.
 Figure 1. Engineering a 16-Mb hemizygous chr7q deletion in iPSCs with Cre-loxP recombination. (A) Overview of strategy for engineering chromosome 7qdeletions with classic Cre-loxP technology. In the first targeting step, a positive selection marker (Puro), a negative selection marker (HSV-tk), and a loxP site were inserted in chr7q (7q36.3 region) in a normal iPSC line using AAV-mediated gene targeting. In the second step, a second loxP site was inserted (7q34 region) in the same orientation together with a different positive selection gene (Neo). In the third step, Cre recombinase was transiently expressed, and clones with chr7q deletions were negatively selected with ganciclovir. IDLV=integrase-deficient lentiviral vector. (B) Upper panel: Schematic of the genomic locus targeted with the Neo-loxP cassette (ATG of the AGK gene). Lower panel: PCR with primers shown in the upper panel amplifying the 5′ and 3′ junctions between the homology arms and the endogenous sequence. (C) Chromosome 7-specific aCGH results for AGK1Cre3 clone. The blue coloring indicates deletion (one copy); the red coloring, amplification (three copies); and the white coloring, normal diploid dosage. Lower panel: chromosome 7 ideogram. The box indicates the ∼16-Mb deleted region in chromosome 7q spanning cytobands 7q34–7q36.3 (approximately 141,256,018–157,150,675) (Supplementary Table E2, online only, available at www.exphem.org). (D) Sanger sequencing of the expected recombined sequence on chr7q. The sequence of the loxP site is in black font; the sequence corresponding to the recombination boundary on region chr7q34, in orange font; and the sequence corresponding to the recombined region of chr7q36.3, in blue font.
Figure 1. Engineering a 16-Mb hemizygous chr7q deletion in iPSCs with Cre-loxP recombination. (A) Overview of strategy for engineering chromosome 7qdeletions with classic Cre-loxP technology. In the first targeting step, a positive selection marker (Puro), a negative selection marker (HSV-tk), and a loxP site were inserted in chr7q (7q36.3 region) in a normal iPSC line using AAV-mediated gene targeting. In the second step, a second loxP site was inserted (7q34 region) in the same orientation together with a different positive selection gene (Neo). In the third step, Cre recombinase was transiently expressed, and clones with chr7q deletions were negatively selected with ganciclovir. IDLV=integrase-deficient lentiviral vector. (B) Upper panel: Schematic of the genomic locus targeted with the Neo-loxP cassette (ATG of the AGK gene). Lower panel: PCR with primers shown in the upper panel amplifying the 5′ and 3′ junctions between the homology arms and the endogenous sequence. (C) Chromosome 7-specific aCGH results for AGK1Cre3 clone. The blue coloring indicates deletion (one copy); the red coloring, amplification (three copies); and the white coloring, normal diploid dosage. Lower panel: chromosome 7 ideogram. The box indicates the ∼16-Mb deleted region in chromosome 7q spanning cytobands 7q34–7q36.3 (approximately 141,256,018–157,150,675) (Supplementary Table E2, online only, available at www.exphem.org). (D) Sanger sequencing of the expected recombined sequence on chr7q. The sequence of the loxP site is in black font; the sequence corresponding to the recombination boundary on region chr7q34, in orange font; and the sequence corresponding to the recombined region of chr7q36.3, in blue font.Engineering hemizygous chr7q deletions up to 22 Mb in normal iPSCs using the CRISPR-Cas9 system
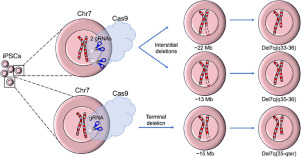
As illustrated in the example described previously, a Cre-loxP strategy can be used to successfully engineer precise deletions of megabase scale. However, it requires multiple successive targeting, drug selection, and single-cell cloning steps, introducing dramatic population bottlenecks and thereby increasing the chances that karyotypically abnormal clones are selected. We therefore next used the CRISPR/Cas9 system in combination with negative selection (HSV-tk) to engineer deletions in only two steps, eliminating the need for pre-insertion of loxP target sites into the genome (Figure 2A). We designed two pairs of gRNAs to engineer targeted deletions 7q35-36.3 and 7q33-36.3 (Supplementary Figure E2 A, online only, available at www.exphem.org). After transduction of the N-2.12-D line (with HSV-tk inserted in 7q36.3) with two lentiviral vectors expressing Cas9 and gRNA targeting 7q35 and 7q36.3 or 7q33 and 7q36.3, ganciclovir-resistant clones were screened by qPCR, probing different regions along the length of chromosome 7. Of 17 clones that received gRNAs targeting 7q35 and 7q36.3, 1 clone was found to harbor a 7q35-qter deletion (clone C(35-qter)13), 1 clone had a 7q35-36.3 deletion (clone C(q35-36)8), 4 clones did not have any detectable 7q deletion, and 11 clones had deletion of the entire chr7 long arm. Of 15 clones that received gRNAs targeting 7q33 and 7q36.3, 1 clone (clone C(q33-36)5) was found to have the predicted 7q33-36.3 deletion, 4 clones did not have any 7q deletion, and 10 had deletion of the entire chr7 long arm. The three different deletion lengths in clones C(35-qter)13, C(q35-36)8, and C(q33-36)5 were confirmed by aCGH (Figure 2B) and by sequencing of the junctions (Supplementary Figure E2 B,C). All three clones were found to be karyotypically normal (Figure 2C). All clones maintained the deletions after 20 passages and had undetectable expression of Cas9 (Supplementary Figure E3, online only, available at www.exphem.org).
 Figure 2. Engineering hemizygous megabase-scale chr7q deletions in normal iPSCs with the CRISPR/Cas9 system. (A) Overview of strategy for engineering chromosome 7q deletions with the CRISPR/Cas9 system. The first step is the same as illustrated in Figure 1A and involves insertion of an HSV-tk transgene in a near-telomeric region of chr7q (7q36.3) for subsequent negative selection. Next, Cas9 and two gRNAs targeting the desired deletion boundaries are delivered through a lentiviral vector [15], and clones with deletions encompassing the location of the HSV-tk transgene are selected with ganciclovir. (B) Chromosome 7-specific aCGH results for the indicated clones (upper panels), and chromosome 7 ideograms with the corresponding deletions annotated (lower panels). Clone C(35-qter)13 harbors an ∼15- Mb deletion spanning nucleotides 144 075,887–159,128,530. Clone C(35–36)8 harbors an ∼13-Mb deletion spanning nucleotides 144,075,887–157,202,735 approximately. Clone C(33–36)5 harbors an ∼22-Mb deletion spanning nucleotides 135,406,029–157,244,681 approximately (Supplementary Table E2). (C)Karyotypic analysis of the indicated del7q-iPSC clones exhibiting normal karyotype (46, XY).
Figure 2. Engineering hemizygous megabase-scale chr7q deletions in normal iPSCs with the CRISPR/Cas9 system. (A) Overview of strategy for engineering chromosome 7q deletions with the CRISPR/Cas9 system. The first step is the same as illustrated in Figure 1A and involves insertion of an HSV-tk transgene in a near-telomeric region of chr7q (7q36.3) for subsequent negative selection. Next, Cas9 and two gRNAs targeting the desired deletion boundaries are delivered through a lentiviral vector [15], and clones with deletions encompassing the location of the HSV-tk transgene are selected with ganciclovir. (B) Chromosome 7-specific aCGH results for the indicated clones (upper panels), and chromosome 7 ideograms with the corresponding deletions annotated (lower panels). Clone C(35-qter)13 harbors an ∼15- Mb deletion spanning nucleotides 144 075,887–159,128,530. Clone C(35–36)8 harbors an ∼13-Mb deletion spanning nucleotides 144,075,887–157,202,735 approximately. Clone C(33–36)5 harbors an ∼22-Mb deletion spanning nucleotides 135,406,029–157,244,681 approximately (Supplementary Table E2). (C)Karyotypic analysis of the indicated del7q-iPSC clones exhibiting normal karyotype (46, XY).Phenotypic characterization of iPSC clones with various deletions of chr7q upon hematopoietic differentiation
We previously found that del7q-iPSCs derived from MDS patients with del7q have severely impaired ability to generate early CD45+ hematopoietic progenitors upon directed in vitro differentiation and that this phenotype is recapitulated by engineered deletions spanning the region 7q32.3-36.1 [4,14]. We thus characterized the hematopoietic differentiation ability of the new panel of clones generated here with partial deletions of this region. Clones C(35-qter)13 and C(35–36)8 generated CD45+ hematopoietic progenitor cells on day 14 of embryoid body-based differentiation with efficiency comparable to that of the parental line (N-2.12-D) (Figure 3A,B). In contrast, clone C(33–36)5, harboring the larger deletion 7q33-7q36.3 exhibited impaired hematopoietic differentiation potential (Figure 3A,B). Colony formation assays in methylcellulose again revealed impaired differentiation ability of the C(33–36)5, but not the other two del7q clones (Figure 3C). These results, taken together with our previous study, allow us to further narrow down our critical region from the previous 20-Mb region to a new 12-Mb region spanning cytobands 7q32.3–7q35 (Figure 3D).
 Figure 3. Hematopoietic phenotypes of CRISPR-Cas9 engineered iPSC clones with chr7q deletions. (A) Representative flow cytometry panels of the indicated clones and the N-2.12-D iPSC line at day 14 of hematopoietic differentiation. (B) Fraction of CD45+ cells on day 14 of hematopoietic differentiation in the indicated del7q clones and the parental normal N-2.12-D line. The mean and SEM of values from four to eight independent differentiation experiments for each clone are shown. (C) Methylcellulose assays at day 14 of hematopoietic differentiation. The number of colonies from 5,000 seeded cells is shown. (D)Upper panel: Schematic of chr7q deletions in the three del7q engineered clones showing the length of the deletions in blue. Lower panel: Chromosome 7 ideogram. The purple box indicates the ∼20-Mb region functionally mapped in our previous study [4]. The red box denotes the overlap of this previously defined 20-Mb region with the region deleted in clone C(33–36)5, which exhibited impaired hematopoietic differentiation, but not in the other two clones, whose hematopoietic differentiation potential was unimpaired. This red box now defines a new smaller critical region of ∼12 Mb spanning cytobands q32.3–q35 (nucleotides 131,706,336–144,075,887 approximately).
Figure 3. Hematopoietic phenotypes of CRISPR-Cas9 engineered iPSC clones with chr7q deletions. (A) Representative flow cytometry panels of the indicated clones and the N-2.12-D iPSC line at day 14 of hematopoietic differentiation. (B) Fraction of CD45+ cells on day 14 of hematopoietic differentiation in the indicated del7q clones and the parental normal N-2.12-D line. The mean and SEM of values from four to eight independent differentiation experiments for each clone are shown. (C) Methylcellulose assays at day 14 of hematopoietic differentiation. The number of colonies from 5,000 seeded cells is shown. (D)Upper panel: Schematic of chr7q deletions in the three del7q engineered clones showing the length of the deletions in blue. Lower panel: Chromosome 7 ideogram. The purple box indicates the ∼20-Mb region functionally mapped in our previous study [4]. The red box denotes the overlap of this previously defined 20-Mb region with the region deleted in clone C(33–36)5, which exhibited impaired hematopoietic differentiation, but not in the other two clones, whose hematopoietic differentiation potential was unimpaired. This red box now defines a new smaller critical region of ∼12 Mb spanning cytobands q32.3–q35 (nucleotides 131,706,336–144,075,887 approximately).Discussion
Here we have reported for the first time the generation of targeted megabase-scale deletions in normal human cells using both the Cre-loxP and CRISPR/Cas9 systems. We also have reported how, in principle, engineering deletions in human iPSCs offer the opportunity to study the phenotypic consequences of specific deletions in relevant cell types generated through directed differentiation.
While the method we describe here allowed us to retrieve the desired clones with predefined deletions, this process is still relatively inefficient and produced a limited number of clones. Notably, a large fraction of clones contained losses of the entire long arm of chr7. We observed this phenomenon in both the Cre-loxP and CRISPR/Cas9 strategies, as well as in our previous inverted loxP site strategy. Thus, these events appear to represent random chromosome losses caused by mitotic errors that are selected in our case because they confer ganciclovir resistance. One can envision future modifications to further improve the efficiency of engineering CRISPR/Cas9-mediated deletions. For example, for interstitial deletions, the simultaneous delivery of a “chimeric” donor template with homology to sequences proximal to the proximal breakpoint and distal to the distal breakpoint could function as a “bridge” to enhance joining of the distal chromosome fragment to the main chromosome through homology-directed repair. On the other hand, a donor DNA with homology to the proximal fragment and containing telomeric sequences could function as a mini-“pseudotelomere” and boost the efficiency of obtaining terminal deletions. Either of these donors could further contain a selectable marker driven by a heterologous promoter or, preferably, by an endogenous locus promoter (“promoter trap”). Another possible and more simple way to improve efficiency could be the simultaneous delivery of more than one gRNA per breakpoint.
As we were able to retrieve and phenotypically study only one clone per deletion, effects of off-target events and/or interclonal variability on the phenotype cannot formally be excluded. Nevertheless, our newly mapped region contains several genes that we and others have previously implicated in del7q-MDS, such as LUC7L2 and HIPK2 [4,17]. Other chr7q genes proposed to play a role in del7q-MDS, including CUX1, SAMD9L, MLL3, and EZH2, as well as the ATP6V0E2 gene that we previously reported, fall outside this 7q32.3–7q35 region [4,18, 19, 20]. It is also possible that combined haplo-insufficiency of more than one gene and/or additional genetic mechanisms are implicated in del7q-MDS that our approach is not optimized to uncover.
The methods we describe are generally applicable and could be used to model a broad range of chromosomal deletions, as well as to functionally characterize regulatory regions of the human genome [21]. Chromosomal deletions of uncertain biological significance are common in both cancer and normal genomes [22,23]. There is evidence that some recurrent deletions drive tumors through cumulative haplo-insufficiencies of more than one genes 24, 25, 26. For the majority, their contributions to the phenotypes of malignant cells are not defined and the driver gene(s) remain elusive. Some deletions may be neutral, occurring in the context of more complex karyotypic abnormalities or marking unstable genomic regions, such as fragile sites [23]. Chromosomal deletions are also found in normal genomes, both in the germline or, more often, occurring de novo and exhibiting varying degrees of mosaicism.
The ability to study deletions with functional assays could provide important information about disease pathogenesis and help identify therapeutic targets.
Acknowledgments
This work was supported by U.S. National Institutes of Health Grant R01HL121570 and by a Scholar Award from the Leukemia and Lymphoma Society to EPP.
Conflict of interest disclosure
The authors declare no competing financial interests.