1. Introduction
Cytochrome P450 monooxygenases (CYPs or P450s) utilize dioxygen and two units of reducing and proton equivalents to catalyze the exemplary monooxygenation reactions. This superfamily of tetrapyrrole heme-thiolate enzymes have the unparalleled ability to catalyze a vast variety of oxidative, peroxidative and reductive reactions such as C H bond hydroxylation, C=C bond epoxidation, heteroatom oxygenation, and many uncommon transformations (Lamb and Waterman, 2013; Dubey and Shaik, 2019; Winkler et al., 2018). The multifunctional CYPs with great catalytic versatility and broad substrate scope are regarded as promising biocatalytic targets, and play essential roles in biotechnological, chemical and pharmaceutical applications, despite the challenges associated with this enzyme superfamily (Lamb and Waterman, 2013; Bernhardt and Urlacher; 2014, Wei et al., 2018). CYPs mediate pivotal steps in primary metabolic pathways and extensively participate in biosynthesis of diverse secondary metabolites (Zhang and Li, 2017). Remarkably, CYPs also act as a means of supplying higher organisms with essential molecules (e.g., sex hormones, brain neurotransmitters, etc.) and play a vital role in protecting the biosystems (Dubey and Shaik, 2019). The promiscuous CYPs involved in the oxidation of diverse substrates display high levels of regio- and stereoselectivity (Šrejber et al., 2018; Munro et al., 2013). Interestingly, in many cases a single amino acid change can alter the CYP's reactivity, selectivity and catalytic efficiency of closely related enzymes (Sezutsu et al., 2013). Based on the global substrate specificity, CYP enzymes fall into two major groups (Urban et al., 2018): (a) the CYPs showing strong preference for a single substrate and (b) the CYPs with loose substrate specificity. Classical examples of the former group include most of the microbial and plant CYPs along with some mammalian CYPs involved in steroidogenesisand eicosanoid biosynthetic pathways, while the latter one encompasses the mammalian CYPs involved in drug/xenobiotic metabolism (Girvan and Munro, 2016; Bernhardt, 2006).
H bond hydroxylation, C=C bond epoxidation, heteroatom oxygenation, and many uncommon transformations (Lamb and Waterman, 2013; Dubey and Shaik, 2019; Winkler et al., 2018). The multifunctional CYPs with great catalytic versatility and broad substrate scope are regarded as promising biocatalytic targets, and play essential roles in biotechnological, chemical and pharmaceutical applications, despite the challenges associated with this enzyme superfamily (Lamb and Waterman, 2013; Bernhardt and Urlacher; 2014, Wei et al., 2018). CYPs mediate pivotal steps in primary metabolic pathways and extensively participate in biosynthesis of diverse secondary metabolites (Zhang and Li, 2017). Remarkably, CYPs also act as a means of supplying higher organisms with essential molecules (e.g., sex hormones, brain neurotransmitters, etc.) and play a vital role in protecting the biosystems (Dubey and Shaik, 2019). The promiscuous CYPs involved in the oxidation of diverse substrates display high levels of regio- and stereoselectivity (Šrejber et al., 2018; Munro et al., 2013). Interestingly, in many cases a single amino acid change can alter the CYP's reactivity, selectivity and catalytic efficiency of closely related enzymes (Sezutsu et al., 2013). Based on the global substrate specificity, CYP enzymes fall into two major groups (Urban et al., 2018): (a) the CYPs showing strong preference for a single substrate and (b) the CYPs with loose substrate specificity. Classical examples of the former group include most of the microbial and plant CYPs along with some mammalian CYPs involved in steroidogenesisand eicosanoid biosynthetic pathways, while the latter one encompasses the mammalian CYPs involved in drug/xenobiotic metabolism (Girvan and Munro, 2016; Bernhardt, 2006).
As CYPs are ubiquitous enzymes with roots in all the eukaryotic taxa, it is too abundant and intricate to elaborate all the systems in one review. Thus, herein, we comprehensively summarize the diversity, versatility and complexity of major eukaryotic CYPs with a special focus on fungal, plant and human CYP systems. This review is aimed to provide a rationalized and consolidated analysis of the membrane-bound eukaryotic CYPs, and the recent conceptual transformation of redox partners from auxiliary electron transfer proteins to CYP functional modulators or regulating partners. Importantly, we elaborate the critical parameters/factors involved in heterologous expression with some recent representative examples and pay special attention on the surrogate cell factories that have enabled successful expression and functional studies of eukaryotic CYPs. The focal point of this review is to address the challenges associated with the heterologous expression of eukaryotic CYPs.
2. Diversity of eukaryotic CYP systems
Evolutionarily, the CYP genes were originated several billion years ago, and are presumed to have prevailed in the last universal common ancestor(Sezutsu et al., 2013). CYPs therefore possess a conserved structural fold despite a considerable variation in their primary sequences. The ubiquitous CYPs are present in all biological kingdoms, and the explosive genome sequencingprojects have identified several hundred thousands of distinct CYP genes (Nelson et al., 2013). However, there are some exceptions since a subgroup of anaerobic microorganisms and microaerophiles lack CYPs (e.g., Escherichia coli). The CYP numbers of eukaryotic organisms range from as low as two genes in yeast (e.g., Schizosaccharomyces pombe) to as many as several hundred genes in plants (e.g., 455 in Oryza sativa and 272 in Arabidopsis thaliana) (Nelson et al., 2013; Urlacher and Girhard, 2012). It is evident that CYPs may not be an essential component of prokaryotes, but it is vital for the emergence of eukaryotic cells in the ancient world of prokaryotic life as sterol biosynthesis(an essential constituent of the plasma membrane) depends on the essential CYP-catalyzed reaction, namely, sterol 14α demethylation by CYP51 (Sezutsu et al., 2013; Omura, 2013). CYPs have not only played a key role in the origin of ancestral monocellular eukaryotic organisms, but also significantly contributed to diversification of eukaryotes. Adaptation of various living organisms to constant environmental changes has resulted in the divergent evolution of CYPs among different taxa, thereby causing significant differences in their catalytic activities and physiological functions (Omura, 2018). The multifunctional eukaryotic CYPs participate in primary and secondary metabolisms by playing key roles in drug/xenobiotic metabolism in humans, and natural product biosynthesis in fungi and plants (Dubey and Shaik, 2019; Bernhardt and Urlacher, 2014; Urlacher and Girhard, 2019). The major physiological functions of eukaryotic CYPs are also related to their essential roles in the synthesis and regulation of several metabolites including the phytohormones (plants), molting and juvenile hormones (insects), as well as the steroid and peptide hormones (vertebrates).
During the long course of evolution, the CYPs of prokaryotes and eukaryotes have differentiated remarkably, and as a result there are no universally conserved residues except the heme-iron coordinating cysteine across the superfamily. CYPs encompass diverse gene families with a perplexing complexity within and between species. Consequently, the numbers of CYP gene families differ a lot among different taxa – bacteria (591 CYP families), fungi (805 CYP families), plants (277 CYP families), insects (208 CYP families), mammals (18 CYP families), archaea (14 CYP families), and viruses (6 CYP families), which reflects the substantial sequence and functional diversity of CYPs (Durairaj et al., 2016; Nelson, 2018). The number of newly recognized CYP sequences has been exponentially increasing with > 300,000 candidates until 2018 (Nelson, 2018), with a possibility of reaching more than one million by 2025 through the Genome 10 K, i5k and GIGA (Global Invertebrate Genomics Alliance) sequencing projects. Though the CYP families share relatively low sequence similarities, the common CYP fold is well conserved (Šrejber et al., 2018). Hitherto, nomenclature has been assigned for > 41,000 CYPs, which includes about 16,000 plant CYPs, 8,000 fungal CYPs, 3,000 bacterial CYPs, and 2,500 mammalian CYPs (Urlacher and Girhard, 2019; Nelson, 2018).
3. Eukaryotic CYP catalytic cycle
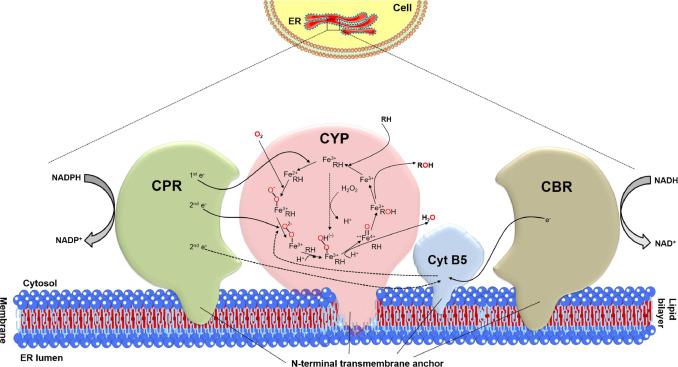
CYPs irrespective of prokaryotic or eukaryotic origin catalyze the prototypical monooxygenation reaction with a common equation as follow: RH + O2 + 2 e− + 2 H+ → ROH + H2O by inserting one oxygen atom of dioxygen into the substrate, while the second oxygen atom is reduced to form a water molecule (Urlacher and Eiben, 2006; McLean et al., 2005). The general catalytic mechanism of a eukaryotic CYP-catalyzed substrate hydroxylation comprises the following sequential steps (Fig. 1): (a) Substrate binding: the substrate (RH) gains access to the active site and binds with the oxidized state of heme iron (Fe3+) by displacing a water molecule over the heme iron; (b) First reduction: the major electron transfer (ET) partner, cytochrome P450 reductase (CPR) transfers the first electron to the ferric heme iron (Fe3+) and forms the reducing ferrous dioxy (Fe2+–O2) complex upon binding of molecular oxygen. Herein, cytochrome b5 (Cyt B5) is not in a position to donate the first electron, as it cannot overcome the redox potential barrier (Barnaba et al., 2017); (c) Second reduction: In this rate-determining step, the second electron can be transferred either from the CPR or Cyt B5, together with the first protonation leading to a super-nucleophilic ferric hydroperoxy (Fe3+–OOH, Compound 0) complex; (d) Oxygen cleavage: heterolytic O–O bond cleavage, the second protonation along with the concurrent loss of a H2O molecule leads to the formation of the highly reactive oxyferryl porphyrin π-cation radical (Fe4+=O, Compound I); (e) Product formation: The hydroxylated product (ROH) is formed upon the abstraction of a hydrogen atom by Compound I and the following OH rebound; and (f) Product release: the monooxygenated product is finally dissociated from the active site, while the enzyme restores its initial ferric state (Fe3+–OH2) and is ready to react again. An alternative route, the peroxide shunt pathway involves the direct binding of H2O2 to the ferric heme iron without the need of O2, NADPH, and redox partner(s) (Zhang and Li, 2017; Munro et al., 2013; Podust and Sherman, 2012).
 Fig. 1. Eukaryotic CYP catalytic cycle and the electron transfer mechanism of CPR/Cyt B5/CB5R redox system.
Fig. 1. Eukaryotic CYP catalytic cycle and the electron transfer mechanism of CPR/Cyt B5/CB5R redox system.4. Heterogeneity of eukaryotic CYP electron transport pathways
To execute the above-described catalytic cycle (Fig. 1), CYPs recruit diversified redox partners to shuttle reducing equivalents. According to the composition of the protein components involved in electron transfer (Fig. 2), CYP systems are categorized into ten distinct classes, of which the eukaryotic CYPs fall into Class I mitochondrial: NADPH→[AdR]→[Adx]→[P450], Class II microsomal A: NADPH→[CPR]→[P450], Class II microsomal B: NADPH→[CPR]→[Cyt B5]→[P450], Class II microsomal C: NADH→[CBR]→[Cyt B5] →[P450] (CBR: NADH-dependent cytochrome b5 reductase), Class VIII: NADPH→[CPR-P450], Class IX: NADH→[P450], and Class X: [P450] systems (Hannemann et al., 2007).
 Fig. 2. Diversity of eukaryotic CYP systems based on the topology of protein components. A. Class I (mitochondrial: NADPH→[AdR]→[Adx]→[P450]); B. Class II microsomal A: NADPH→[CPR]→[P450]; C. Class II microsomal B: NADPH→[CPR]→[Cyt B5] →[P450]; D. Class II microsomal C: NADH→[CBR]→[Cyt B5]→[P450]; E. Class VIII (NADPH→[CPR-P450]); F. Class IX (NADH→[P450]); and G. Class X [P450] systems.
Fig. 2. Diversity of eukaryotic CYP systems based on the topology of protein components. A. Class I (mitochondrial: NADPH→[AdR]→[Adx]→[P450]); B. Class II microsomal A: NADPH→[CPR]→[P450]; C. Class II microsomal B: NADPH→[CPR]→[Cyt B5] →[P450]; D. Class II microsomal C: NADH→[CBR]→[Cyt B5]→[P450]; E. Class VIII (NADPH→[CPR-P450]); F. Class IX (NADH→[P450]); and G. Class X [P450] systems.Class I system, the most common prokaryotic (bacterial) P450 system, is also present in certain eukaryotes especially mammals, but not in plants and fungi. It is hypothesized that the microsomal CYP is the ancestor of mammalian mitochondrial CYP system, wherein the ER-targeting sequence at the amino-terminus of microsomal CYP was transformed to a mitochondria-targeting sequence possibly by accumulated point mutations during the course of evolution (Omura and Gotoh, 2017). Eukaryotic Class I mitochondrial P450 system consists of a membrane-bound CYP and a reducing system comprising two components, adrenodoxin (Adx) located in the mitochondrial matrix and NADPH-dependent adrenodoxin reductase (AdR) bound to the inner mitochondrial membrane (Fig. 2A). Most of eukaryotic CYPs (especially fungal, plants and mammals) adopt a Class II system and catalyze extremely diverse reactions in terms of both reaction types and substrate scope. Class II microsomal A system is the most common in eukaryotes, which comprises two integral membrane proteins, namely, CYP and CPR. The CPR is responsible for the sequential transfer of reducing equivalents (two electrons) from NAD(P)H to the heme-iron via the prosthetic cofactors FAD and FMN (Fig. 2B). Class II microsomal B system recruits a third auxiliary protein, Cyt B5, to transfer the second electron to the oxyferrous CYP (Fig. 2C). Class II microsomal C system encompasses Cyt B5 and CBR, where the electrons are directly transferred to certain CYPs without the need of CPR (Ichinose and Wariishi, 2012; Stiborová et al., 2016) (Fig. 2D). In addition to the abundant Class II system, some fungal species also encompass Class VIII and Class IX systems. In Class VIII system, the N-terminal heme domain is naturally fused to a C-terminal CPR domain through a short peptide linker (P450foxy) (Shoun and Takaya, 2002), in which the electrons are supplied in an intra-molecular manner (Fig. 2E). Thus, this class of CYPs that are mainly originated from fungi are catalytically self-sufficient. Class IX has an exceptional single-component system (e.g., P450nor), whereby CYP is able to accept electrons directly from NAD(P)H without the requirement of any additional redox partners (Shoun and Takaya, 2002). Class IX is functionally different from all other CYPs because the soluble P450nor mediates denitrification in fungi by catalyzing the reduction of two molecules of NO to form N2O (Fig. 2F). Remarkably, while most CYP systems depend on redox partner(s) for ET, P450s belonging to Class X system neither require molecular oxygen nor any electron source for their catalysis (Fig. 2G). Class X CYPs capable of using an independent intramolecular ET system have been reported in some plant (e.g., CYP74A-D) and mammalian (e.g., CYP5A1 and CYP8A1) species (Hannemann et al., 2007).
5. Significance of N-terminal transmembrane domain in eukaryotic CYP systems
The eukaryotic CYPs along with its counterparts are characteristically attached on the cytoplasmic side of endoplasmic reticulum (ER) or the matrix side of the inner mitochondrial membrane. With the progress of modern biotechnology, a better understanding of eukaryotic CYPs and its association with membranes has been emerging. The eukaryotic CYPs which were once considered to be integral membrane protein with several transmembrane segments, were recently determined to be membrane-associated proteins with a short N-terminal membrane anchoring domain (Šrejber et al., 2018). To date, the knowledge about these membrane-bound CYPs has been assembled into a defined model in course of progression and attracts growing attention. The new model depicts that the eukaryotic CYPs are anchored to the ER through an N-terminal transmembrane α-helix with the N-terminus lying on the luminal side, while the catalytic domain of CYP reclines on the cytosolic side (Fig. 1) (Šrejber et al., 2018; Urban et al., 2018). The CYP catalytic domain is slightly submerged in the lipid bilayer with its proximal side facing cytosol, whereas the N-terminus and F/G loop lying on the distal side of the enzyme are deeply immersed. Apparently, the signal peptide sequence of N-terminal anchor governs the trafficking of CYPs into ER or mitochondria (Šrejber et al., 2018). The phospholipid composition in the membrane-bound CYP systems is crucial for protein folding and stability, whilst plays an essential role in the ET required for CYP monooxygenation (Barnaba et al., 2017). Of note, the composition of membrane influences the function of CYP, as the amino acid composition and configuration of N-terminal helix varies significantly in different CYP families (Šrejber et al., 2018; Gideon et al., 2012). The N-terminal transmembrane domain (TMD) is mainly composed of 20-30 hydrophobic amino acid residues which facilitates the interaction with hydrophobic ER membrane environment.
Though the primarily role of TMD is to mediate association to the lipid membrane, it also plays an essential role in the interaction between CYP and CPR, substrate binding and other downstream catalytic steps (Maroutsos et al., 2019).The major diflavin reductase, CPR carrying both FAD and FMN is also attached to the membrane similar to CYP via an N-terminal membrane-binding domain (Fig. 1) (Mukherjee et al., 2021). A typical microsomal CYP system forms a membrane-bound protein–protein complex through the electrostatic interactions between the positively charged residues on the proximal side of CYP and negatively charged amino acids of the CPR's FMN domain (Šrejber et al., 2018; Scott et al., 2016). The membrane binding domain significantly contributes to the efficient electron transfer from CPR to CYP, and the interactions correspond to the oxidation state of CPR and binding of cofactor (Mukherjee et al., 2021; Xia et al., 2019). The membrane environment thus plays a crucial role in mediating the formation of binary complex and influences the interaction/cooperation of integral CPR-CYP electron mediating system (Gideon et al., 2012). The active site of eukaryotic CYP is profoundly concealed within the structure, and connected to the protein surface and exterior by multiplex access/egress channels. As the heme is tilted towards the membrane surface, the binding and positioning of CYPs on membranes is vigorous and catalytically relevant. It has to be noted that the charge of phospholipid bilayeris likely to alter the orientation of the CYP catalytic domain relative to the membrane. The orientation of CYPs in the phospholipid bilayer could facilitate ease in substrate access and product egress channels as the B/C and F/G loops are pointed beneath the phospholipid head groups, while the solvent channel is positioned toward the membrane-water interface (Urban et al., 2018; Berka et al., 2013). In membrane-bound eukaryotic CYPs, the substrate binds to the active site of CYP (substrate binding pocket) by entering through cytosol or membrane interior resulting in possible substrate/product trafficking, whereas the substrate access and product egress are directly mediated by the same channel in bacterial CYPs.
While most of the prokaryotic CYPs are soluble, almost all the membrane-bound eukaryotic CYPs are insoluble upon bacterial expression which hinders the functional and structural studies (Denisov et al., 2012; Zelasko et al., 2013). Several eukaryotic CYPs of fungal, plant and human origins were reported to be expressed in bacterial systems mostly upon N-terminal sequence changes via deletions or modifications. Of note, the multifunctional eukaryotic CYP protein structures are still under-represented in the Protein Data Bank(https://www.rcsb.org/) owing to the difficulties associated with the membrane-bound nature. Indeed, except for the two full-length microsomal CYPs, viz., CYP51 lanosterol 14-α demethylase of Saccharomyces cerevisiae (PDB ID: 4LXJ and 5EQB) (Monk et al., 2014) and CYP19 aromatase of Homo sapiens(PDB ID: 3EQM) (Ghosh et al., 2009), the TMDs are almost always cleaved in other structures to obtain complete solubilisation and subsequent crystallization. The structures obtained through N-terminal cleavage or deletion are though biochemically relevant, the reported results are devoid of knowledge relating to the structure, topology or molecular organization of these membrane-bound eukaryotic CYPs (Šrejber et al., 2018; Barnaba et al., 2017). It is worth mentioning that the truncation of TMD might not only affect the CYP's phospholipid composition leading to folding and stability issues, but could also influence the CYP-CPR interactions and impair the catalytic functionality.
6. Surrogate cell factories for eukaryotic CYP production
Recombinant expression of membrane-bound eukaryotic CYPs in surrogate (mainly microbial) cell factories provides a cost-effective avenue for large scale protein production for downstream biochemical and structural studies. Nevertheless, functional expression and production of eukaryotic CYPs in surrogate cell factories is not simple and entails some fundamental problems (Durairaj et al., 2019). Some of the vital parameters for the heterologous recombinant expression of eukaryotic CYPs include (a) selection of host strain, (b) choice of the expression plasmid with an appropriate promoter, (c) codon optimization of exons, (d) culture conditions, (e) protein induction and so on (Table 1). It has become the foremost priority to identify an appropriate surrogate cell factory for functional expression of eukaryotic CYPs, since handling such metalloenzymes could be rather tricky owing to their membrane-bound property, low expression level, protein misfolding, ineffective substrate uptake, and need as well as tolerance for rich redox cofactors. In order to facilitate sustained expression of membrane-bound eukaryotic CYPs, a wide range of prokaryotic and eukaryotic surrogate cell factories have been successfully developed (Table 1). Herein, we will focus on the pros and cons of these microbial cell factories, as well as the established and arising engineering strategies/approaches for eukaryotic CYP production. Though the scope of this review is primarily focused on microbial cell factories, a brief overview on the higher-eukaryotic cell factories (viz., plant and mammalian hosts) are also included in order to provide a holistic understanding on eukaryotic CYP expression.
Table 1. Holistic representation of expression of eukaryotic membrane-bound cytochrome P450s in different surrogate cell factories.
| Parameters | Heterologous expression of eukaryotic membrane-bound CYPs | ||||
|---|---|---|---|---|---|
| Bacterial system | Yeast system | Fungal system | Plant system | Mammalian cell-line system | |
| Selection of vector, promoter and host strain | Required | Required | Required | Required | Required |
| Optimization of culture conditions | Required | Optional | Optional | Optional | Optional |
| Duration of culture for production | Fast (several hours to a day) | Moderate (several days) | Moderate (several days to a week) | Slow (several days to weeks) | Slow (several weeks) |
| Culture cost & technical demand | Low | Low | Moderate | High | High |
| Supplementation of heme precursor 5-aminolevulinic acid | Required | Optional | Not required | Not required | Not required |
| Optimization of codon usage | Required | Required | Required | Optional | Optional |
|
Reduction of secondary mRNA structure |
Required | Optional | Optional | Optional | Optional |
| N-terminal modification | Deletion of N-TMD or construction of chimeric CYPs is required | Not necessary | Not necessary | Not necessary | Not necessary |
| Post-translational modifications | Does not occur | Offers higher eukaryotic-style post-translational modifications | Offers higher eukaryotic-style post-translational modifications | Yes | Yes |
| Presence of endogenous CYPs | No | Very few | Several to many endogenous CYPs | Several to many endogenous CYPs | Several to many endogenous CYPs |
| Co-expression of redox partner | Required | Co-expression of CPR is advantageous | Optional | Optional | Optional |
| Co-expression of auxiliary proteins | Chaperones are required for proper folding and high protein expression | Optional | Optional | Not required | Not required |
| C-terminal fusion | C-terminal GFP-based platforms allow increased CYP expression and fluorescence report | Not required | Not required | Not required | Not required |
| Protein purification & structural studies | Highly purified CYP proteins can be achieved, allowing structural studies | Microsomes can be purified upon tedious efforts, but structural studies are limited | Hard to achieve | Hard to achieve | Hard to achieve |
| Biochemical characterization | Purified enzymes | Whole-cell biotransformation is preferred over purified enzymes or microsomal fractions | Whole-cell biotransformation is preferred over purified enzymes or microsomal fractions | Whole-cell biotransformation is preferred over purified enzymes | Microsomal fractions |
6.1. Conventional microbial cell factory systems
The most conventional and preferred surrogate cell factories for the heterologous expression of CYPs are the bacterial and yeast host systems. Several studies concerning the heterologous expression of membrane-bound eukaryotic CYPs in the traditional cell factory systems (E. coli and S. cerevisiae) have been well documented (Winkler et al., 2018; Zelasko et al., 2013; Freigassner et al., 2009; Sørensen and Mortensen, 2005; Yun et al., 2006; Ichinose and Wariishi, 2013; Emmerstorfer et al., 2014; Ichinose et al., 2015; Faiq et al., 2014; Stiborová et al., 2017; Hausjell et al., 2018; Hausjell et al., 2020; Park et al., 2020).
6.1.1. Bacterial cell factory as eukaryotic CYP production platform
Escherichia coli has always been the first-choice bacterial cell factory for heterologous CYP expression due to its unparalleled fast growth kinetics, inexpensive and rich complex media, high cell density cultures, and ease of genetic manipulation. With the extensive knowledge on genetics and physiology, many state-of-the-art molecular tools and protocols have successfully been developed for recombinant protein production and purification, facilitating functional and structural studies (Table 1). Besides, the absence of native CYP genes in E. coli makes it ideal for recombinant CYP production due to the advantage of lacking cross-interference. Owing to these advantages, a wide range of CYP-mediated biotransformations have been demonstrated in this bacterial cell factory leading to milligram- to gram-scale production of various compounds (Zelasko et al., 2013; Yun et al., 2006; Ichinose et al., 2015; Ajikumar et al., 2010). However, several factors concerning the prototypical elements of eukaryotic CYPs viz. the presence of N-terminal hydrophobic TMD comprising the conserved proline-rich residues prior to the CYP catalytic domain, demand for the addition of the precursor for heme prosthetic group, and dependency on ET proteins as well as NAD(P)H cofactor for electron supply complicate the heterologous CYP expression (Fig. 3). Remarkably, the major drawback with the bacterial cell factory is that this system often demands the membrane-bound eukaryotic CYPs to undergo significant modifications (mutagenesis/deletions) to obtain soluble fractions, possibly resulting in serious consequences. N-terminal CYP modifications though sporadically resolves the problems, the alterations could influence the native enzyme's interaction and fail the desired catalytic activity. Besides, this approach has turned out to be case-specific (not successful for all membrane-bound CYPs), since several N-terminal altered CYPs were still expressed as membrane fractions due to their indelible hydrophobic topologies. Though various strategies have been constructively developed (discussed in detail in Section 7), there is still no a universal guideline to enable successful eukaryotic CYP overexpression. The success rate varies upon a case-by-case basis, and therefore optimizations have to be performed discretely for every membrane-bound CYP protein.
 Fig. 3. Challenges associated with the prokaryotic and eukaryotic expression of membrane-bound eukaryotic CYPs.
Fig. 3. Challenges associated with the prokaryotic and eukaryotic expression of membrane-bound eukaryotic CYPs.6.1.2. Yeast cell factory as eukaryotic CYP production platform
Saccharomyces cerevisiae, the eukaryotic counterpart of E. coli serves as an ideal cell factory due to its salient features such as genetic accessibility, rapid growth, and convenient genetic and metabolic engineering approaches. The conventional yeast system is desirable for eukaryotic CYP expression as it offers rich ER environment in combination with the higher eukaryotic protein synthesis machinery, thereby enabling the recombinant expression of full-length membrane-bound CYPs without any genetic modifications/truncations (Table 1). S. cerevisiae offers an innate intracellular heme environment with the presence of three well characterized CYP enzymes including CYP51, CYP56 and CYP61, thereby providing an in vivo self-sufficient background for the heterologous CYPs ensuring complete saturation by hemin prosthetic group. The endogenous CYP environment of yeast cell factory is a significant attribute that lacks in E. coli, where supplementation of expensive heme precursors (5-aminolevuleic acid) is often demanded for the up-regulation of CYP activity. Besides, the added advantage of presence of native S. cerevisiae CPR can directly supply reducing equivalents to the heterologous CYPs, wherein an additional step of recombinant CPR co-expression is eliminated, thus avoiding ancillary stress to the host organism leading to lower CYP production. Another interesting attribute of yeast cell factory is its ability to regenerate the cofactor NADPH intracellularly since the effective coupling between NADPH and CYP is crucial for large-scale applications involving high substrate concentration(Park et al., 2020). Of note, the yeast cell factory allows biocatalytic investigations of recombinant CYPs through a one-pot biotransformation procedure achieved by either growing or resting (non-growing) cells (Durairaj et al., 2016; Lundemo and Woodley, 2015). Alternatively, the recombinant CYPs can be studied by yeast microsomes, favoring in vitro work concerning kinetics and reaction mechanism. Nevertheless, though yeast microsomal experiments are more precise, the factors such as sophisticated isolation process, culture-to-culture variations, and technical constraints in purification make it a complicated and cumbersome process to acquire active functional enzymes (Durairaj et al., 2016; Drăgan et al., 2011; Yan et al., 2017). As a result, yeast whole-cell biotransformation is preferred for CYP functional characterizations owing to its simplicity, straightforward analysis, and biocatalyst stability (Durairaj et al., 2016; Park et al., 2020; Lundemo and Woodley, 2015); though the kinetic and structural investigations are limited. The major drawback with the yeast biotransformation is the protein/product yield especially with the strains expressing heterologous CYPs (Fig. 3). Though gram-scale production is possible in yeast-based fermentations or whole-cell biotransformations, the range of production titer often varies and demands longer incubation (Park et al., 2020). Another limitation of yeast cell factory is that certain substrates and/or products may face some permeabilization issues, as the charged molecules are less likely to permeate multiple cell membranes (Yan et al., 2017).
6.1.3. Amalgamation of conventional bacterial and yeast cell factory platform
On the whole, our current knowledge on eukaryotic CYPs results from the composition of researches on both bacterial and yeast cell factories. On one hand, the bacterial cell factory aids higher yield of eukaryotic CYPs (14–20 nmol/mg protein), although the N-terminal modifications are inevitable. On the other hand, though the yield is relatively low (∼1 nmol/mg protein), yeast cell factory enables expression of membrane-bound CYPs in its native form without any modifications (Hausjell et al., 2018). The space-time yield analysis of several CYP biotransformations in bacterial and yeast cell factories has demonstrated that though the production titer can be comparable (1-2 g/L/h), the yeast system demands longer incubation (100-200 h reaction time) compared to that of bacterial system (Park et al., 2020). Of note, the CYP activity might differ depending upon the surrogate cell factory employed in terms of not only expression rate but also functionality. For instance, the recombinant human CYP2S1 expressed in E. coli and S. cerevisiae systems gave varied results as the enzyme's catalytic specificity was altered (Wu et al., 2006, Nishida et al., 2010). Thus, direct or combinational comparison of full-length membrane-bound CYPs in both the bacterial and yeast expression systems is required to enable a better functional and mechanistic understanding. Interestingly, microbial production of plant benzylisoquinoline alkaloids were successfully established in an combinatory co-culture system (Minami et al., 2008). The transgenic E. coli cells expressing the branchpoint intermediate (S-reticuline) biosynthetic gene were co-cultured with the S. cerevisiae cells expressing CYP80G2 along with other required biosynthetic genes (coclaurine-N-methyltransferase/berberine bridge enzyme). This co-culture system efficiently produced magnoflorine and scoulerine with a final yield of 7.2 and 8.3 mg/L, respectively. Likewise, metabolic pathway distribution within microbial consortium could solve the technical difficulties and significantly enhance the production of natural products. Incorporation of robust production of taxadiene in E. coli and the functional expression of CYP enzyme taxadiene 5α-hydroxylase in S. cerevisiae resulted in the highest production (33 mg/L yield) of oxygenated taxanes (Zhou et al., 2015). Amalgamation of both bacterial and yeast based CYP expression could therefore be a novel approach to solve the limitations and exploit the advantages of each expression system.
6.2. Unconventional microbial cell factory system
Unconventional (non-Saccharomyces) yeasts such as fission yeastSchizosaccharomyces pombe (Bureik et al., 2002), methylotrophic yeast Pichia pastoris (Gudiminchi et al., 2013), dimorphic yeasts Yarrowia lipolytica(Mauersberger et al., 2013) and Arxula adeninivorans (Theron et al., 2014), lactose-utilizing yeast Kluyveromyces lactis (Engler et al., 2000) and its thermophilic sister strain Kluyveromyces marxianus (Engler et al., 2000) can also be employed as alternate surrogate cell factory systems for heterologous CYP production. Of which, P. pastoris and Y. lipolytica have been proven effective for the recombinant production of several membrane-bound CYP proteins with higher yields (Iwama et al., 2016; Garrigós-Martínez et al., 2021). Recently, in order to investigate whether baker yeast is a superior host to express membrane-bound CYPs, direct comparison on the expression of full-length chalcone 3-hydroxylase, a CYP essential in the flavonoid pathway was expressed in three different strains of P. pastoris and S. cerevisiae (Hausjell et al., 2020). Regarding productivity it has been reported that a highest yield of 600 pmol/mg protein was achieved in controlled bioreactor cultivations using the P. pastorisstrain KM71H; while the yield of S. cerevisiae is twice as low as the lowest product yield obtained in P. pastoris strain SMD1168H. Interestingly, S. pombewhich is an underestimated cell factory for CYP studies was recently demonstrated to be an ideal surrogate cell factory for expression and functional characterization of several hard-to-express and so-called orphan human CYPs including CYP4Z1, CYP2A7, CYP4A22 and CYP20A1 (Durairaj et al., 2019; Yan et al., 2017; Bureik et al., 2002; Durairaj et al., 2019; Durairaj et al., 2020). A classic example on heterologous expression and recombinant production of CYP2C9 in three different cell factories including the baculovirus transfected insect cell lines, E. coli, and S. pombe towards the hydroxylation of non-steroidal anti-inflammatory drug diclofenac resulted in the preparation of 2.2 mg, 110 mg and 2.8 g of 4’-hydroxy metabolite representing an overall yield of 28 %, 35 % and 75 %, respectively (Winkler et al., 2018; Drăgan et al., 2011; Rushmore et al., 2000; Vail et al., 2005). Besides, several human drug-metabolizing enzymes (Winkler et al., 2018) including CYP2D6, CYP2C9, CYP3A4, CYP11B1, CYP11B2 and CYP21 were successfully expressed in S. pombe, and the kinetic parameterswere determined using the whole-cell biocatalysts. Recently, a functional library of human CYPome was constructed in S. pombe, where all the 57 human CYPs were co-expressed along with their natural human ET (CPR/AdR+Adx) partners (Durairaj et al., 2019). This complete library of recombinant fission yeast strains demonstrated functional expression of unmodified sequences of human PAN CYPome, and their catalytic activity were determined within a single surrogate cell factory for the first time.
6.3. Higher-eukaryotic cell factory systems
6.3.1. Fungal cell factory as eukaryotic CYP production platform
An evolutionarily closer organism, a filamentous fungus could serve as a preferred cell factory for the expression of homologous and heterologous membrane-bound eukaryotic CYPs (Durairaj et al., 2016; Zhang et al., 2021). Filamentous fungi could overcome several limitations faced in traditional expression systems (bacteria / yeast), including mRNA precursor maturation, higher splicing-rate, efficient protein secretion machinery and post-translational modifications (Tanaka et al., 2014; Nevalainen and Peterson, 2014). With the advent of modern biotechnology, several species of filamentous fungi have emerged as promising cell factories for the production of pharmaceutically relevant proteins with salient improvements. Of which, Aspergillus spp. (Meyer et al., 2011) dominate the scene as desirable expression hosts for eukaryotic CYP production as many studies have been reported with Aspergillus nidulans (Liu et al., 2019), Aspergillus oryzae (Cao et al., 2019), Aspergillus niger(Faber et al., 2001), Aspergillus sojae (Araki et al., 2019) and Aspergillus aculeatus(Thiele et al., 2020). Several fungal CYPs involved in the natural product biosynthetic pathways were functionally elucidated using the fungal cell factory with A. oryzae or A. nidulans as heterologous host (Zhang et al., 2021). For instance, the biosynthetic pathways of helvolic acid (Lv et al., 2017), fusidic acid(Cao et al., 2019), and cephalosporin P1 (Cao et al., 2019) from the ascomycetous fungi Acremonium fumigatus, A. fusidioides, and A. chrysogenum were studied using A. oryzae as an expression host system. Another example demonstrated the functional investigation of biosynthetic genes of fungal meroterpenoid from the filamentous fungus Acremonium egyptiacum in Aspergillus spp., wherein the genes ascA-D involved in the biosynthesis of ascofuranone and ascochlorin were expressed in A. oryzae, while AscE-G proteins were expressed in A. sojae high-copy expression system (Araki et al., 2019). Of which, AscE is a soluble CYP/reductase fusion protein that catalyzes stereoselective epoxidation of the terminal double bond of the prenyl group, while AscF (terpene cyclase) and AscG (CYP) are membrane-bound proteins involved in the terpene cyclizationand oxidation of ilicicolin into ascochlorin. Based on the preliminary experimental evidence it is suggested that the heterologous production of fungal CYPs yields better results in fungal cell factory than CYPs of other origins. Recombinant production of the active form of mammalian CYP proteins in fungal cell factory is often limited by the differences in the mode of glycosylation of mammalian and fungal cells. Filamentous fungal system features the high-mannose type of glycosylation, but lacks the mammalian-style terminal sialylation of glycans which may affect the functionality, serum half-life and immunogenicity of recombinant proteins (Nevalainen and Peterson, 2014). In addition, factors such as DNA manipulation, incorrect processing / misfolding and secretory yields may also add up to the list of limitations associated with fungal cell factory for recombinant CYP production (Tanaka et al., 2014; Nevalainen and Peterson, 2014).
6.3.2. Plant cell factory as eukaryotic CYP production platform
Plant CYPs involved in secondary metabolism are sometimes difficult to express in a microbial cell factory, and therefore several plant-based hosts have been developed to procure sufficient protein expression and improve the yield of desired compounds. Plant-based heterologous expression offers several advantages as they permit defined mRNA and protein processing, protein subcellular localization and metabolic compartmentalization, and have essential metabolic precursors and coenzymes (Table 1). Though plant (N- and O-linked) and mammalian (terminal sialylation of glycans) cells feature different glycosylation patterns, the variances do not impair the recombinant protein production; and tremendous efforts have been made for humanization of protein N-glycosylation in plant cell factory (Gomord et al., 2010). A wild relative of tobacco, Nicotiana benthamiana, serving as an efficient production system of flu vaccines at industrial-scale (Marsian and Lomonossoff, 2016), has also acted as a competent cell factory for the heterologous expression of several plant CYPs (Reed et al., 2017). The multifunctional enzyme AsCYP51H10 involved in the modification of C and D rings of the pentacyclic triterpene scaffold into 12,13β-epoxy-3β,16β-dihydroxy-oleanane, was successfully expressed and studied using this transient plant expression system (Geisler et al., 2013). Moreover, co-expression of additional heterologous redox partner is not necessary since the N. benthamiana CPR provides sufficient electron equivalents to mediate CYP catalysis. Recently, the members of CYP79C family belonging to the glucosinolate (GLS) biosynthetic pathway in Arabidopsis thaliana were functionally characterized through Agrobacterium-mediated transient expression in N. benthamiana (Wang et al., 2020). Functional investigation of CYP79C1 and CYP79C2 in the GLS pathway engineered N. benthamiana facilitated simultaneous testing of substrate specificity against multiple aliphatic and aromatic amino acids. The whole-genome sequenced non-vascular plant Physcomitrella patens offered efficient homologous recombination, and facilitated recombinant production of several commercially important pharmaceutical proteins (Khairul Ikram et al., 2017). Furthermore, the cinnamic acid 4-hydroxylase from the aquatic plant Anthoceros agrestis,which could not be expressed in yeast system possibly due to high GC contentand/or different codon usage, was successfully expressed in the haploid plant Physcomitrella patens and functionally characterized at biochemical and molecular levels (Wohl and Petersen, 2020). Specifically, A. thaliana serves as an excellent model for recombinant protein production and has been extensively studied for the developmental and molecular biology, as well as pharmaceutical applications (Von Schaewen et al., 2018). Remarkably, the recently developed Arabidopsis-based recombinant protein production platform (Jeong et al., 2018) is expected to serve as an efficient cell factory for the heterologous CYP production suitable for biochemical and structural studies. Interestingly, the plant cell factory system offers efficient endogenous redox partner system (e.g., N. benthamiana CPR) that can effectively pair up with the heterologous CYPs for direct ET. However, the rich intracellular heme environment with enormous native/endogenous CYPs may also have a downside due to the cross-link effect and/or interference with the desired recombinant CYP protein of interest. Though there are successful instances, functional studies in plant cell factories remain limited with early-stage objectives due to the restrictions including enzyme stability, relatively low turn-over, yield, tedious protein extraction and purification process, and high cost of downstream processing (Schillberg et al., 2019).
6.3.3. Mammalian cell-line factory as eukaryotic CYP production platform
Development of genetically modified mammalian cell lines to functionally express membrane-bound CYPs represents another successful strategy by facilitating practical in vitro approaches for enzymatic characterization, drug metabolism screening and early detection of drug toxicity (Xuan et al., 2016; Satoh et al., 2017) (Table 1). The cell-line factory allows recombinant production of larger and complex proteins including membrane-bound CYPs as it offers inherent transcriptional and translational environment along with appropriate chaperonin, secretory pathway and redox assembly coupled with efficient protein folding, and excellent post-translational modifications. Though the glycosylation pattern of some of the cell lines slightly varies to that of human-type glycosylations; the cell lines can be fine-tuned by codon optimization including glycoengineering which improves the efficacy and enhances the recombinant expression (Hunter et al., 2019). Alternatively, human cell line (e.g., HEK-293) systems represent an ideal source for CYP mediated drug metabolism studies, and allow post-translational modifications of membrane-bound eukaryotic proteins for functional production at high levels. Cell lines can be readily transfected or virally transduced, and the recombinant CYP proteins can be produced either transiently or by stable expression (Hunter et al., 2019). Primary human hepatocytes, hepatic cell lines, and stem cell derived hepatocytes are primarily utilized as in vitro models for the recombinant CYP production and functional studies. Nevertheless, high donor to donor variability, scarcity, limited lifespan, low expression and yield restrict the efficient utilization of major cell lines for enzymatic analysis involving CYP mediated drug metabolism. Currently, several alternative cell line platforms have been developed, and tremendous efforts have been put forward to determine CYP functions with the advances of synthetic biology and next-generation engineering to achieve gram-scale productivity (Hunter et al., 2019; Gutiérrez-González et al., 2019; Boon et al., 2020). For instance, human hepatoma celllines can demonstrate increased stability, ample life-span and accessibility compared to the traditional cell-based assays using primary human hepatocytes. Transchromosomic HepG2 cell lines have facilitated expression of four major CYPs (CYP2C9, 2C19, 2D6, and 3A4) and CPR using the mammalian-derived artificial chromosome vector (Satoh et al., 2017). The expression levels were significantly higher than that of the parental HepG2 cells demonstrating a highly versatile model for the evaluation of drug-drug interactions and screening of hepatotoxicity. Another interesting study demonstrated expression of 14 human CYPs in HepG2-derived cell lines individually with the aid of lentiviral expression system (Xuan et al., 2016). In order to determine the most suitable expression platform for the in vitro CYP enzymatic activities, four different mammalian cell lines (COS-7, HepG2, 293T and 293FT) were functionally investigated with the typical variants of CYP2C9, CYP2C19 and CYP2D6 (Dai et al., 2015). The results indicated that the fast-growing variant of 293 cell line 293FT demonstrated higher levels of expression as well as in vitroactivities. Recently, another interesting study successfully developed an ideal heterologous expression system with three CYP isoforms (CYP1A2, CYP2C9, and CYP3A4) using mammalian 293FT cells by optimizing high-precision conditions (Kumondai et al., 2020). Therein, the highest CYP expression which can be quantifiable by CO-difference spectroscopy was achieved in a lost-cost manner by replacing expensive transfection reagent with cost-effective and efficient substitute PEI-Max, and demonstrated significantly higher enzymatic activity by co-expressing CPR and Cyt B5. Though the mammalian cell line factory serves as an excellent system for recombinant human CYP production and in vitro functional studies, some issues concerning expression, reduced activity and low CYP inducibility remain to be addressed. Besides, factors such as demand for expensive culture media, complex growth requirements, technical demand, time-consuming procedures, lengthy expression phase, lot-to-lot heterogeneity, and scalability are some of the major bottlenecks to be overcome.
7. Approaches to eukaryotic CYP production in surrogate cell factories
In order to explore the functional and structural features of eukaryotic CYPs, suitable quantities of purified, well-folded and catalytically active proteins are required. The membrane-bound eukaryotic CYPs display higher complexity than most of soluble prokaryotic CYPs (Denisov et al., 2012). Recombinant production of eukaryotic CYPs remains a challenging task owing to the constraints in obtaining sufficient soluble proteins due to non-expression, protein misfolding, or aggregation into insoluble inclusion bodies(Durairaj et al., 2016; Zelasko et al., 2013). Bacterial system though serves as an ideal cell factory for prokaryotic CYPs, the membrane-bound nature of the eukaryotic CYPs often impedes their heterologous expression and recombinant production. As discussed above, membrane-bound proteins behave poorly in overexpression systems and tend to be unstable in the detergent solutions that are used in the membrane extraction and purification steps. Consequently, tremendous efforts have been undertaken in optimizing target proteins, diverse expression systems and purification strategies to yield sufficient proteins (Durairaj et al., 2016; Maroutsos et al., 2019; Zelasko et al., 2013; Yun et al., 2006; Ichinose et al., 2015; Hausjell et al., 2018). Despite great endeavors on optimizing the overexpression of membrane-bound CYPs, there still lacks a systematic approach with a solid theoretical basis. Instead, optimizations can be achieved only via a tedious trial-and-error process (Durairaj et al., 2016). Herein, we elucidate some of the key strategies that favor efficient and effective expression and production of membrane-bound eukaryotic CYPs in surrogate cell factories.
7.1. Codon optimization
With the advent of modern biotechnology, gene synthesis with adapted codon usage has become a convenient strategy to ensure sustained production of membrane-bound proteins in heterologous systems (Claassens et al., 2017). For the recombinant protein production, the codons are optimized according to the expression host system by harmonizing the favorable GC-content, and by excluding the mRNA secondary structures at the 5’ untranslated region (Fig. 4). Besides the unfavorable motifs including the repeats, Shine-Dalgarno like sequences and RNase sites also need to be excluded in order to promote heterologous protein expression. The expression levels of several mammalian and plant CYPs were significantly improved in bacterial cell factories upon codon optimization (Yamaguchi et al., 2021; Wu et al., 2009). Furthermore, transcriptional tuning and usage of rare codons has proven successful for membrane-bound proteins as it slows down the translation and allows proper co-translational folding of specific domains (α-helices and β-sheets) and insertion of membrane (Claassens et al., 2017). This not only facilitates appropriate translocation rates but also ensures membrane integration thereby avoiding accumulation of inclusion bodies.